
Sewage-associated gemycircularvirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus sewopo3; Sewage derived gemycircularvirus 3
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
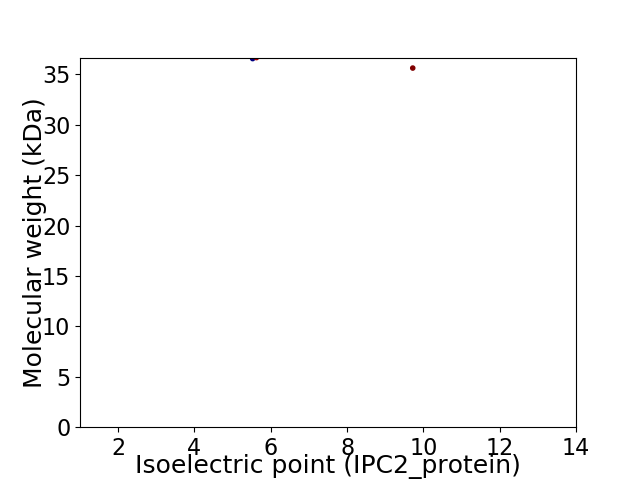
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7CL87|A0A0A7CL87_9VIRU Replication-associated protein OS=Sewage-associated gemycircularvirus 6 OX=1519401 PE=3 SV=1
MM1 pKa = 7.62SSFSFHH7 pKa = 6.37ARR9 pKa = 11.84YY10 pKa = 9.75ALLTYY15 pKa = 8.93AQCGDD20 pKa = 4.24LCPFTIVDD28 pKa = 4.65LLSTMGAEE36 pKa = 4.28CIIGRR41 pKa = 11.84EE42 pKa = 4.01HH43 pKa = 6.42HH44 pKa = 6.33QDD46 pKa = 3.17GGIHH50 pKa = 5.88LHH52 pKa = 5.67VFVDD56 pKa = 4.46FGRR59 pKa = 11.84KK60 pKa = 7.79YY61 pKa = 10.01RR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84ADD67 pKa = 3.3TFDD70 pKa = 3.1VGGFHH75 pKa = 7.34PNISQSYY82 pKa = 6.35GTPEE86 pKa = 3.51KK87 pKa = 10.88GYY89 pKa = 10.89DD90 pKa = 3.65YY91 pKa = 11.08ACKK94 pKa = 10.67DD95 pKa = 3.3GDD97 pKa = 4.13VVAGGLDD104 pKa = 3.18RR105 pKa = 11.84PVPTEE110 pKa = 3.67GRR112 pKa = 11.84GGDD115 pKa = 3.77SKK117 pKa = 9.73THH119 pKa = 6.28SIWSQITSATTRR131 pKa = 11.84EE132 pKa = 4.34SFWDD136 pKa = 3.95LVHH139 pKa = 7.71DD140 pKa = 5.62LDD142 pKa = 4.59PKK144 pKa = 10.56SAVTCFTQLQKK155 pKa = 11.25YY156 pKa = 9.43CDD158 pKa = 3.1WKK160 pKa = 11.12YY161 pKa = 10.72RR162 pKa = 11.84YY163 pKa = 9.41CPPAYY168 pKa = 9.27EE169 pKa = 4.48SPAGARR175 pKa = 11.84FRR177 pKa = 11.84NDD179 pKa = 2.89TSDD182 pKa = 4.64GRR184 pKa = 11.84GDD186 pKa = 3.27WLLQSGIGGARR197 pKa = 11.84VGRR200 pKa = 11.84VKK202 pKa = 10.87SLVLYY207 pKa = 10.59GPSQTGKK214 pKa = 6.66TTWARR219 pKa = 11.84SLGAHH224 pKa = 7.37IYY226 pKa = 9.89QVGLLSGSEE235 pKa = 4.17CMKK238 pKa = 11.07APDD241 pKa = 3.17VEE243 pKa = 4.49YY244 pKa = 10.88AVFDD248 pKa = 5.02DD249 pKa = 4.07IRR251 pKa = 11.84GGMKK255 pKa = 9.69FFPSFKK261 pKa = 9.92EE262 pKa = 3.7WLGCQPHH269 pKa = 5.71VCVKK273 pKa = 10.06EE274 pKa = 4.09LYY276 pKa = 9.83RR277 pKa = 11.84EE278 pKa = 3.93PRR280 pKa = 11.84VIEE283 pKa = 3.94WGKK286 pKa = 8.54PAIWCSNADD295 pKa = 3.41PRR297 pKa = 11.84DD298 pKa = 3.73DD299 pKa = 3.64MSYY302 pKa = 11.47CDD304 pKa = 4.3VQWMEE309 pKa = 4.25ANCTFIEE316 pKa = 4.19ITEE319 pKa = 4.35KK320 pKa = 11.12LLDD323 pKa = 3.58WEE325 pKa = 4.44
MM1 pKa = 7.62SSFSFHH7 pKa = 6.37ARR9 pKa = 11.84YY10 pKa = 9.75ALLTYY15 pKa = 8.93AQCGDD20 pKa = 4.24LCPFTIVDD28 pKa = 4.65LLSTMGAEE36 pKa = 4.28CIIGRR41 pKa = 11.84EE42 pKa = 4.01HH43 pKa = 6.42HH44 pKa = 6.33QDD46 pKa = 3.17GGIHH50 pKa = 5.88LHH52 pKa = 5.67VFVDD56 pKa = 4.46FGRR59 pKa = 11.84KK60 pKa = 7.79YY61 pKa = 10.01RR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84ADD67 pKa = 3.3TFDD70 pKa = 3.1VGGFHH75 pKa = 7.34PNISQSYY82 pKa = 6.35GTPEE86 pKa = 3.51KK87 pKa = 10.88GYY89 pKa = 10.89DD90 pKa = 3.65YY91 pKa = 11.08ACKK94 pKa = 10.67DD95 pKa = 3.3GDD97 pKa = 4.13VVAGGLDD104 pKa = 3.18RR105 pKa = 11.84PVPTEE110 pKa = 3.67GRR112 pKa = 11.84GGDD115 pKa = 3.77SKK117 pKa = 9.73THH119 pKa = 6.28SIWSQITSATTRR131 pKa = 11.84EE132 pKa = 4.34SFWDD136 pKa = 3.95LVHH139 pKa = 7.71DD140 pKa = 5.62LDD142 pKa = 4.59PKK144 pKa = 10.56SAVTCFTQLQKK155 pKa = 11.25YY156 pKa = 9.43CDD158 pKa = 3.1WKK160 pKa = 11.12YY161 pKa = 10.72RR162 pKa = 11.84YY163 pKa = 9.41CPPAYY168 pKa = 9.27EE169 pKa = 4.48SPAGARR175 pKa = 11.84FRR177 pKa = 11.84NDD179 pKa = 2.89TSDD182 pKa = 4.64GRR184 pKa = 11.84GDD186 pKa = 3.27WLLQSGIGGARR197 pKa = 11.84VGRR200 pKa = 11.84VKK202 pKa = 10.87SLVLYY207 pKa = 10.59GPSQTGKK214 pKa = 6.66TTWARR219 pKa = 11.84SLGAHH224 pKa = 7.37IYY226 pKa = 9.89QVGLLSGSEE235 pKa = 4.17CMKK238 pKa = 11.07APDD241 pKa = 3.17VEE243 pKa = 4.49YY244 pKa = 10.88AVFDD248 pKa = 5.02DD249 pKa = 4.07IRR251 pKa = 11.84GGMKK255 pKa = 9.69FFPSFKK261 pKa = 9.92EE262 pKa = 3.7WLGCQPHH269 pKa = 5.71VCVKK273 pKa = 10.06EE274 pKa = 4.09LYY276 pKa = 9.83RR277 pKa = 11.84EE278 pKa = 3.93PRR280 pKa = 11.84VIEE283 pKa = 3.94WGKK286 pKa = 8.54PAIWCSNADD295 pKa = 3.41PRR297 pKa = 11.84DD298 pKa = 3.73DD299 pKa = 3.64MSYY302 pKa = 11.47CDD304 pKa = 4.3VQWMEE309 pKa = 4.25ANCTFIEE316 pKa = 4.19ITEE319 pKa = 4.35KK320 pKa = 11.12LLDD323 pKa = 3.58WEE325 pKa = 4.44
Molecular weight: 36.56 kDa
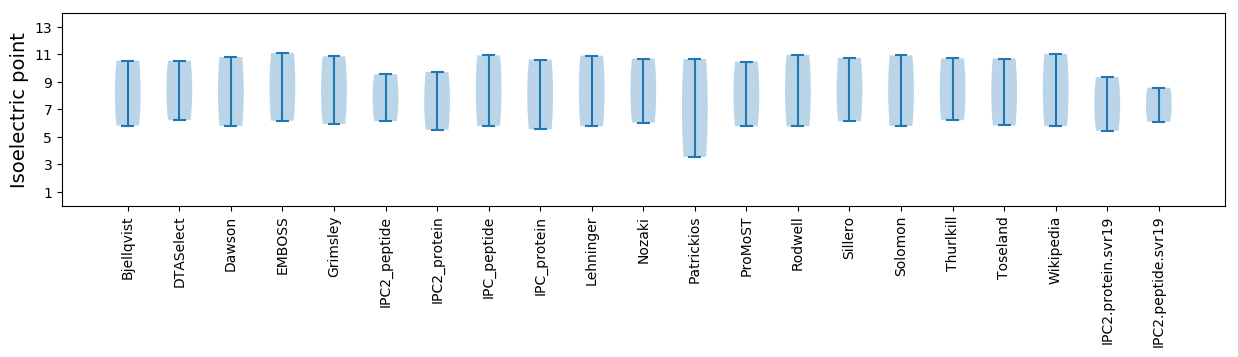
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7CL87|A0A0A7CL87_9VIRU Replication-associated protein OS=Sewage-associated gemycircularvirus 6 OX=1519401 PE=3 SV=1
MM1 pKa = 7.02THH3 pKa = 7.41PSFICQQPNMIRR15 pKa = 11.84SRR17 pKa = 11.84RR18 pKa = 11.84TRR20 pKa = 11.84SRR22 pKa = 11.84RR23 pKa = 11.84SVRR26 pKa = 11.84PRR28 pKa = 11.84RR29 pKa = 11.84PLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ITKK38 pKa = 9.73RR39 pKa = 11.84RR40 pKa = 11.84PTVKK44 pKa = 10.16RR45 pKa = 11.84AISKK49 pKa = 10.22KK50 pKa = 9.82RR51 pKa = 11.84ILNITSRR58 pKa = 11.84KK59 pKa = 9.05KK60 pKa = 10.33RR61 pKa = 11.84DD62 pKa = 3.84VMNQWSNTTADD73 pKa = 4.15GGSKK77 pKa = 10.34PITNGIVTLAGPVGGYY93 pKa = 8.74FYY95 pKa = 10.34WQATARR101 pKa = 11.84TLDD104 pKa = 3.41QGPGQIGIVSDD115 pKa = 3.56EE116 pKa = 4.29ATRR119 pKa = 11.84TATTCFMRR127 pKa = 11.84GLSEE131 pKa = 5.64RR132 pKa = 11.84IDD134 pKa = 3.35IQTNSSLPWRR144 pKa = 11.84WRR146 pKa = 11.84RR147 pKa = 11.84ICITTKK153 pKa = 10.67DD154 pKa = 3.39NSYY157 pKa = 11.23DD158 pKa = 3.72HH159 pKa = 7.37SDD161 pKa = 3.43LGALQAYY168 pKa = 8.22GGWLLSSAGIVRR180 pKa = 11.84PWVNSYY186 pKa = 10.88INNNGTTQAGQWVDD200 pKa = 3.59LFEE203 pKa = 4.87GAGNVDD209 pKa = 3.11WADD212 pKa = 3.52VNSAKK217 pKa = 10.03IDD219 pKa = 3.36TRR221 pKa = 11.84RR222 pKa = 11.84IDD224 pKa = 3.26LHH226 pKa = 6.5YY227 pKa = 11.16DD228 pKa = 2.93KK229 pKa = 10.65TITINSGNSSGVFRR243 pKa = 11.84TYY245 pKa = 10.77RR246 pKa = 11.84RR247 pKa = 11.84WHH249 pKa = 6.37PMNKK253 pKa = 9.52NLVYY257 pKa = 10.86DD258 pKa = 4.63DD259 pKa = 4.9DD260 pKa = 4.58QDD262 pKa = 3.76GGKK265 pKa = 8.88MDD267 pKa = 3.99SSHH270 pKa = 7.38LSVTDD275 pKa = 3.18KK276 pKa = 11.14RR277 pKa = 11.84GMGNYY282 pKa = 8.97IVLDD286 pKa = 3.85FFQPHH291 pKa = 6.51ASATSADD298 pKa = 3.96RR299 pKa = 11.84LILRR303 pKa = 11.84SNSALFWHH311 pKa = 6.89EE312 pKa = 3.9KK313 pKa = 9.73
MM1 pKa = 7.02THH3 pKa = 7.41PSFICQQPNMIRR15 pKa = 11.84SRR17 pKa = 11.84RR18 pKa = 11.84TRR20 pKa = 11.84SRR22 pKa = 11.84RR23 pKa = 11.84SVRR26 pKa = 11.84PRR28 pKa = 11.84RR29 pKa = 11.84PLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ITKK38 pKa = 9.73RR39 pKa = 11.84RR40 pKa = 11.84PTVKK44 pKa = 10.16RR45 pKa = 11.84AISKK49 pKa = 10.22KK50 pKa = 9.82RR51 pKa = 11.84ILNITSRR58 pKa = 11.84KK59 pKa = 9.05KK60 pKa = 10.33RR61 pKa = 11.84DD62 pKa = 3.84VMNQWSNTTADD73 pKa = 4.15GGSKK77 pKa = 10.34PITNGIVTLAGPVGGYY93 pKa = 8.74FYY95 pKa = 10.34WQATARR101 pKa = 11.84TLDD104 pKa = 3.41QGPGQIGIVSDD115 pKa = 3.56EE116 pKa = 4.29ATRR119 pKa = 11.84TATTCFMRR127 pKa = 11.84GLSEE131 pKa = 5.64RR132 pKa = 11.84IDD134 pKa = 3.35IQTNSSLPWRR144 pKa = 11.84WRR146 pKa = 11.84RR147 pKa = 11.84ICITTKK153 pKa = 10.67DD154 pKa = 3.39NSYY157 pKa = 11.23DD158 pKa = 3.72HH159 pKa = 7.37SDD161 pKa = 3.43LGALQAYY168 pKa = 8.22GGWLLSSAGIVRR180 pKa = 11.84PWVNSYY186 pKa = 10.88INNNGTTQAGQWVDD200 pKa = 3.59LFEE203 pKa = 4.87GAGNVDD209 pKa = 3.11WADD212 pKa = 3.52VNSAKK217 pKa = 10.03IDD219 pKa = 3.36TRR221 pKa = 11.84RR222 pKa = 11.84IDD224 pKa = 3.26LHH226 pKa = 6.5YY227 pKa = 11.16DD228 pKa = 2.93KK229 pKa = 10.65TITINSGNSSGVFRR243 pKa = 11.84TYY245 pKa = 10.77RR246 pKa = 11.84RR247 pKa = 11.84WHH249 pKa = 6.37PMNKK253 pKa = 9.52NLVYY257 pKa = 10.86DD258 pKa = 4.63DD259 pKa = 4.9DD260 pKa = 4.58QDD262 pKa = 3.76GGKK265 pKa = 8.88MDD267 pKa = 3.99SSHH270 pKa = 7.38LSVTDD275 pKa = 3.18KK276 pKa = 11.14RR277 pKa = 11.84GMGNYY282 pKa = 8.97IVLDD286 pKa = 3.85FFQPHH291 pKa = 6.51ASATSADD298 pKa = 3.96RR299 pKa = 11.84LILRR303 pKa = 11.84SNSALFWHH311 pKa = 6.89EE312 pKa = 3.9KK313 pKa = 9.73
Molecular weight: 35.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
638 |
313 |
325 |
319.0 |
36.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.956 ± 0.138 | 2.508 ± 1.041 |
7.524 ± 0.332 | 3.135 ± 1.248 |
3.605 ± 0.705 | 8.777 ± 0.746 |
2.665 ± 0.288 | 5.643 ± 0.931 |
4.545 ± 0.049 | 6.113 ± 0.243 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.038 ± 0.134 | 3.605 ± 1.656 |
4.389 ± 0.373 | 3.605 ± 0.154 |
8.621 ± 1.721 | 8.15 ± 0.534 |
6.897 ± 0.947 | 5.329 ± 0.361 |
3.135 ± 0.04 | 3.762 ± 0.596 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
