
Silicimonas algicola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Silicimonas
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
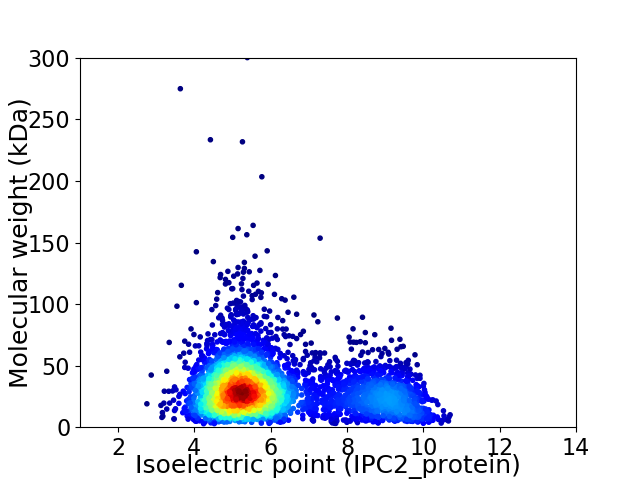
Virtual 2D-PAGE plot for 4315 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316G8Z5|A0A316G8Z5_9RHOB Glutamate 5-kinase OS=Silicimonas algicola OX=1826607 GN=proB PE=3 SV=1
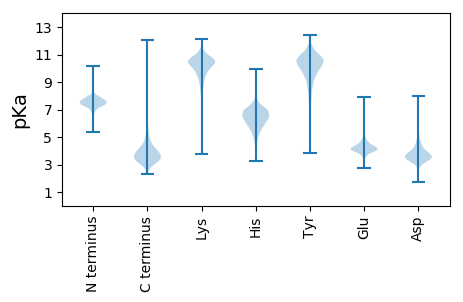
MM1 pKa = 6.96TCKK4 pKa = 10.58ALIPGLLALATLSGCSGGGGGAVYY28 pKa = 10.94ADD30 pKa = 4.14GPSPASIDD38 pKa = 4.03EE39 pKa = 4.23IVGLQFPLRR48 pKa = 11.84SARR51 pKa = 11.84LTSDD55 pKa = 4.85DD56 pKa = 4.52GFTTANAGLSTYY68 pKa = 9.87TIEE71 pKa = 4.34FTSGTTANLTTPAGTVEE88 pKa = 4.12LTDD91 pKa = 3.65GGGMNFTGSLNGSDD105 pKa = 4.24YY106 pKa = 11.15FLTSLGWGHH115 pKa = 7.39DD116 pKa = 3.64FLEE119 pKa = 4.69GLGLEE124 pKa = 4.63VTDD127 pKa = 3.93GMGNVLVATGVFGLEE142 pKa = 4.01TRR144 pKa = 11.84PEE146 pKa = 4.4DD147 pKa = 3.62VPEE150 pKa = 4.22AAVVANFLGGSEE162 pKa = 4.08LHH164 pKa = 6.05GTVNDD169 pKa = 3.5VPFFDD174 pKa = 3.86TGAANLTADD183 pKa = 5.04FGAQTVGGNVFVGGGTTLAFDD204 pKa = 4.24GPVPIVGNTFSGDD217 pKa = 3.59LEE219 pKa = 4.45IFSATATLNEE229 pKa = 4.1GTATGRR235 pKa = 11.84FYY237 pKa = 11.68GDD239 pKa = 3.12AAQVVAGTYY248 pKa = 10.08SGSATEE254 pKa = 4.82AGSDD258 pKa = 3.27VDD260 pKa = 4.04FAGVFHH266 pKa = 7.6ADD268 pKa = 2.94
MM1 pKa = 6.96TCKK4 pKa = 10.58ALIPGLLALATLSGCSGGGGGAVYY28 pKa = 10.94ADD30 pKa = 4.14GPSPASIDD38 pKa = 4.03EE39 pKa = 4.23IVGLQFPLRR48 pKa = 11.84SARR51 pKa = 11.84LTSDD55 pKa = 4.85DD56 pKa = 4.52GFTTANAGLSTYY68 pKa = 9.87TIEE71 pKa = 4.34FTSGTTANLTTPAGTVEE88 pKa = 4.12LTDD91 pKa = 3.65GGGMNFTGSLNGSDD105 pKa = 4.24YY106 pKa = 11.15FLTSLGWGHH115 pKa = 7.39DD116 pKa = 3.64FLEE119 pKa = 4.69GLGLEE124 pKa = 4.63VTDD127 pKa = 3.93GMGNVLVATGVFGLEE142 pKa = 4.01TRR144 pKa = 11.84PEE146 pKa = 4.4DD147 pKa = 3.62VPEE150 pKa = 4.22AAVVANFLGGSEE162 pKa = 4.08LHH164 pKa = 6.05GTVNDD169 pKa = 3.5VPFFDD174 pKa = 3.86TGAANLTADD183 pKa = 5.04FGAQTVGGNVFVGGGTTLAFDD204 pKa = 4.24GPVPIVGNTFSGDD217 pKa = 3.59LEE219 pKa = 4.45IFSATATLNEE229 pKa = 4.1GTATGRR235 pKa = 11.84FYY237 pKa = 11.68GDD239 pKa = 3.12AAQVVAGTYY248 pKa = 10.08SGSATEE254 pKa = 4.82AGSDD258 pKa = 3.27VDD260 pKa = 4.04FAGVFHH266 pKa = 7.6ADD268 pKa = 2.94
Molecular weight: 26.73 kDa
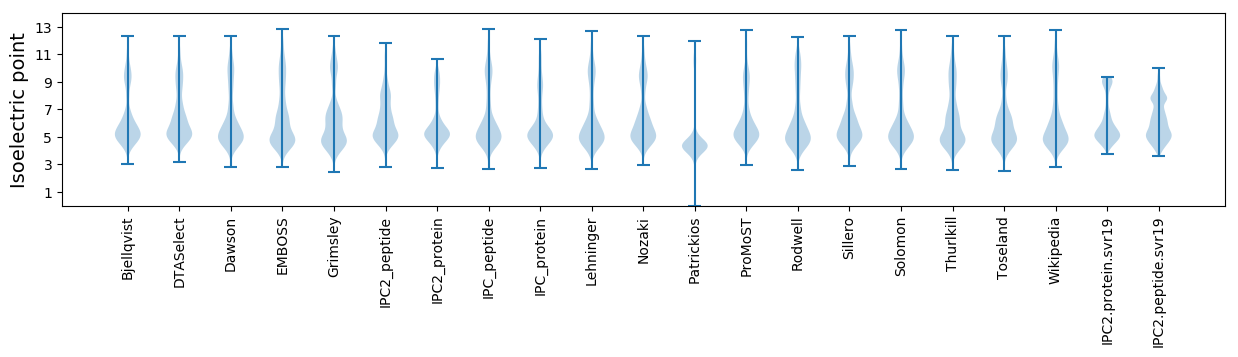
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316GD04|A0A316GD04_9RHOB Uncharacterized protein DUF1446 OS=Silicimonas algicola OX=1826607 GN=C8D95_101634 PE=4 SV=1
MM1 pKa = 7.26LHH3 pKa = 5.01HH4 pKa = 6.98HH5 pKa = 6.47FRR7 pKa = 11.84RR8 pKa = 11.84FLDD11 pKa = 4.25ALDD14 pKa = 5.01DD15 pKa = 4.48LNPAQIEE22 pKa = 4.09DD23 pKa = 4.06AQTKK27 pKa = 9.83IRR29 pKa = 11.84DD30 pKa = 3.36IRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.1TEE36 pKa = 4.47AISEE40 pKa = 4.06IEE42 pKa = 3.7ARR44 pKa = 11.84TNRR47 pKa = 11.84EE48 pKa = 4.06HH49 pKa = 7.25KK50 pKa = 10.76CPFCGDD56 pKa = 3.07EE57 pKa = 5.39RR58 pKa = 11.84RR59 pKa = 11.84QKK61 pKa = 9.63WGRR64 pKa = 11.84TRR66 pKa = 11.84TKK68 pKa = 8.8IQRR71 pKa = 11.84YY72 pKa = 8.2RR73 pKa = 11.84CSGCRR78 pKa = 11.84KK79 pKa = 7.68TYY81 pKa = 10.22SGRR84 pKa = 11.84TGSAIGRR91 pKa = 11.84IHH93 pKa = 7.36RR94 pKa = 11.84PDD96 pKa = 3.69LFLVALRR103 pKa = 11.84DD104 pKa = 3.82MLDD107 pKa = 3.03ASAPQSVRR115 pKa = 11.84KK116 pKa = 9.05LARR119 pKa = 11.84QLDD122 pKa = 4.16LNKK125 pKa = 9.1YY126 pKa = 6.29TVWRR130 pKa = 11.84WRR132 pKa = 11.84MIVFSIIGSRR142 pKa = 11.84SVATSFSGIIEE153 pKa = 4.19ADD155 pKa = 3.05EE156 pKa = 4.5TYY158 pKa = 10.6QRR160 pKa = 11.84EE161 pKa = 4.19SRR163 pKa = 11.84KK164 pKa = 10.06GSRR167 pKa = 11.84EE168 pKa = 3.88WVHH171 pKa = 6.21HH172 pKa = 5.8SRR174 pKa = 11.84SAEE177 pKa = 4.25CYY179 pKa = 9.1SAAAPAVGRR188 pKa = 11.84LHH190 pKa = 7.69DD191 pKa = 4.36ARR193 pKa = 11.84AEE195 pKa = 3.74NDD197 pKa = 3.04ARR199 pKa = 11.84SFEE202 pKa = 4.38VATPILTVADD212 pKa = 4.07RR213 pKa = 11.84SGARR217 pKa = 11.84LFQRR221 pKa = 11.84LPNRR225 pKa = 11.84KK226 pKa = 8.75RR227 pKa = 11.84GTVEE231 pKa = 4.2RR232 pKa = 11.84AMQPLVPGDD241 pKa = 4.1AVLCSDD247 pKa = 4.83GGNGYY252 pKa = 10.43KK253 pKa = 10.19SLAAARR259 pKa = 11.84GLVHH263 pKa = 6.49FVVGSRR269 pKa = 11.84PGTRR273 pKa = 11.84VAAGCYY279 pKa = 9.58HH280 pKa = 6.13IQNVNSLHH288 pKa = 5.65ARR290 pKa = 11.84YY291 pKa = 9.74GKK293 pKa = 10.22FIRR296 pKa = 11.84PFCGPATKK304 pKa = 10.48NLNGYY309 pKa = 8.78IRR311 pKa = 11.84WLEE314 pKa = 3.69VRR316 pKa = 11.84LAGVRR321 pKa = 11.84PAEE324 pKa = 4.29VVRR327 pKa = 11.84ASS329 pKa = 3.1
MM1 pKa = 7.26LHH3 pKa = 5.01HH4 pKa = 6.98HH5 pKa = 6.47FRR7 pKa = 11.84RR8 pKa = 11.84FLDD11 pKa = 4.25ALDD14 pKa = 5.01DD15 pKa = 4.48LNPAQIEE22 pKa = 4.09DD23 pKa = 4.06AQTKK27 pKa = 9.83IRR29 pKa = 11.84DD30 pKa = 3.36IRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.1TEE36 pKa = 4.47AISEE40 pKa = 4.06IEE42 pKa = 3.7ARR44 pKa = 11.84TNRR47 pKa = 11.84EE48 pKa = 4.06HH49 pKa = 7.25KK50 pKa = 10.76CPFCGDD56 pKa = 3.07EE57 pKa = 5.39RR58 pKa = 11.84RR59 pKa = 11.84QKK61 pKa = 9.63WGRR64 pKa = 11.84TRR66 pKa = 11.84TKK68 pKa = 8.8IQRR71 pKa = 11.84YY72 pKa = 8.2RR73 pKa = 11.84CSGCRR78 pKa = 11.84KK79 pKa = 7.68TYY81 pKa = 10.22SGRR84 pKa = 11.84TGSAIGRR91 pKa = 11.84IHH93 pKa = 7.36RR94 pKa = 11.84PDD96 pKa = 3.69LFLVALRR103 pKa = 11.84DD104 pKa = 3.82MLDD107 pKa = 3.03ASAPQSVRR115 pKa = 11.84KK116 pKa = 9.05LARR119 pKa = 11.84QLDD122 pKa = 4.16LNKK125 pKa = 9.1YY126 pKa = 6.29TVWRR130 pKa = 11.84WRR132 pKa = 11.84MIVFSIIGSRR142 pKa = 11.84SVATSFSGIIEE153 pKa = 4.19ADD155 pKa = 3.05EE156 pKa = 4.5TYY158 pKa = 10.6QRR160 pKa = 11.84EE161 pKa = 4.19SRR163 pKa = 11.84KK164 pKa = 10.06GSRR167 pKa = 11.84EE168 pKa = 3.88WVHH171 pKa = 6.21HH172 pKa = 5.8SRR174 pKa = 11.84SAEE177 pKa = 4.25CYY179 pKa = 9.1SAAAPAVGRR188 pKa = 11.84LHH190 pKa = 7.69DD191 pKa = 4.36ARR193 pKa = 11.84AEE195 pKa = 3.74NDD197 pKa = 3.04ARR199 pKa = 11.84SFEE202 pKa = 4.38VATPILTVADD212 pKa = 4.07RR213 pKa = 11.84SGARR217 pKa = 11.84LFQRR221 pKa = 11.84LPNRR225 pKa = 11.84KK226 pKa = 8.75RR227 pKa = 11.84GTVEE231 pKa = 4.2RR232 pKa = 11.84AMQPLVPGDD241 pKa = 4.1AVLCSDD247 pKa = 4.83GGNGYY252 pKa = 10.43KK253 pKa = 10.19SLAAARR259 pKa = 11.84GLVHH263 pKa = 6.49FVVGSRR269 pKa = 11.84PGTRR273 pKa = 11.84VAAGCYY279 pKa = 9.58HH280 pKa = 6.13IQNVNSLHH288 pKa = 5.65ARR290 pKa = 11.84YY291 pKa = 9.74GKK293 pKa = 10.22FIRR296 pKa = 11.84PFCGPATKK304 pKa = 10.48NLNGYY309 pKa = 8.78IRR311 pKa = 11.84WLEE314 pKa = 3.69VRR316 pKa = 11.84LAGVRR321 pKa = 11.84PAEE324 pKa = 4.29VVRR327 pKa = 11.84ASS329 pKa = 3.1
Molecular weight: 37.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1295017 |
25 |
2773 |
300.1 |
32.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.386 ± 0.054 | 0.856 ± 0.012 |
6.189 ± 0.031 | 6.047 ± 0.033 |
3.725 ± 0.025 | 8.829 ± 0.033 |
2.008 ± 0.019 | 4.898 ± 0.027 |
2.712 ± 0.027 | 10.145 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.621 ± 0.018 | 2.277 ± 0.019 |
5.154 ± 0.026 | 2.694 ± 0.023 |
7.366 ± 0.035 | 5.256 ± 0.026 |
5.563 ± 0.025 | 7.699 ± 0.027 |
1.466 ± 0.015 | 2.109 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
