
Rusa timorensis papillomavirus type 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
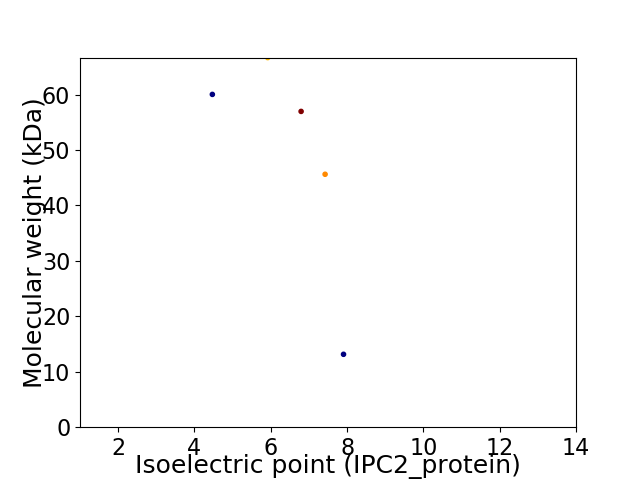
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R2Z1A7|A0A2R2Z1A7_9PAPI Major capsid protein L1 OS=Rusa timorensis papillomavirus type 2 OX=1905556 GN=L1 PE=3 SV=1
MM1 pKa = 7.25FVFLSVCVVTLIMATVNRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.87RR22 pKa = 11.84ASDD25 pKa = 2.84WDD27 pKa = 3.87LYY29 pKa = 9.78RR30 pKa = 11.84ACRR33 pKa = 11.84GGADD37 pKa = 4.17CISDD41 pKa = 3.59VVNKK45 pKa = 10.35YY46 pKa = 6.97EE47 pKa = 4.71HH48 pKa = 6.65KK49 pKa = 9.43TPADD53 pKa = 3.16KK54 pKa = 10.92VLQYY58 pKa = 11.11GGAGVFLGGLGIGTGSGALGVRR80 pKa = 11.84PGVTGPAEE88 pKa = 4.63EE89 pKa = 4.69IPLVTLGEE97 pKa = 4.29RR98 pKa = 11.84TTITDD103 pKa = 3.47TGSGRR108 pKa = 11.84TPSLGSTSFGGTSRR122 pKa = 11.84FGSGFGGRR130 pKa = 11.84IDD132 pKa = 5.37LISAGRR138 pKa = 11.84GRR140 pKa = 11.84VTTSDD145 pKa = 3.3VSVNTVDD152 pKa = 3.67SVTPSVILPEE162 pKa = 4.31TVPADD167 pKa = 3.63PGIEE171 pKa = 3.9LQVFTDD177 pKa = 3.78VEE179 pKa = 4.56DD180 pKa = 3.94SVQVTVSSSVDD191 pKa = 3.23EE192 pKa = 4.86GEE194 pKa = 4.39VAVLEE199 pKa = 4.68VPATEE204 pKa = 4.3SFGRR208 pKa = 11.84PSQVTTRR215 pKa = 11.84GGGGKK220 pKa = 8.06VHH222 pKa = 7.25HH223 pKa = 6.86SRR225 pKa = 11.84HH226 pKa = 4.92VVPAEE231 pKa = 3.67TGTILGEE238 pKa = 4.05TSEE241 pKa = 4.32TQNIFIEE248 pKa = 4.87GSGVGDD254 pKa = 3.87SYY256 pKa = 11.86SEE258 pKa = 4.63SIEE261 pKa = 3.73LWTYY265 pKa = 10.08SRR267 pKa = 11.84PRR269 pKa = 11.84YY270 pKa = 7.31STPDD274 pKa = 2.98SEE276 pKa = 4.71PPVNRR281 pKa = 11.84RR282 pKa = 11.84GINNPFSKK290 pKa = 9.95RR291 pKa = 11.84YY292 pKa = 8.0YY293 pKa = 9.57KK294 pKa = 10.3QVTVEE299 pKa = 4.61DD300 pKa = 4.29PQFLTEE306 pKa = 3.97PQKK309 pKa = 10.92LVSNVFEE316 pKa = 4.5NPAFVDD322 pKa = 4.48DD323 pKa = 4.74VDD325 pKa = 5.37DD326 pKa = 5.62SIPFLDD332 pKa = 4.11TDD334 pKa = 3.55TRR336 pKa = 11.84PYY338 pKa = 9.27NTEE341 pKa = 3.75SDD343 pKa = 3.73FLDD346 pKa = 3.22IGRR349 pKa = 11.84LGRR352 pKa = 11.84VQYY355 pKa = 9.37TVSPGEE361 pKa = 3.84GLGVSRR367 pKa = 11.84IGTRR371 pKa = 11.84FTIKK375 pKa = 9.26TRR377 pKa = 11.84SGVTIGEE384 pKa = 4.35QVHH387 pKa = 5.04YY388 pKa = 9.6RR389 pKa = 11.84YY390 pKa = 9.31PLSPITGASEE400 pKa = 5.24DD401 pKa = 3.88IEE403 pKa = 4.3LQNLVTGMSEE413 pKa = 4.38GVLDD417 pKa = 4.04SLNDD421 pKa = 3.9TVVEE425 pKa = 4.14TLNNAMLEE433 pKa = 4.32SVDD436 pKa = 5.33LDD438 pKa = 4.77DD439 pKa = 5.92ISDD442 pKa = 3.96FLSVSSSVLEE452 pKa = 4.02NEE454 pKa = 4.92LMDD457 pKa = 3.93NVEE460 pKa = 5.53DD461 pKa = 3.57ITFGQLAFRR470 pKa = 11.84DD471 pKa = 3.76EE472 pKa = 4.74GEE474 pKa = 4.57TEE476 pKa = 4.15VFTYY480 pKa = 10.37PDD482 pKa = 3.28YY483 pKa = 11.9SNTQKK488 pKa = 11.22AGVTVISEE496 pKa = 4.49DD497 pKa = 3.4NVTGTKK503 pKa = 10.28SGDD506 pKa = 3.53VVNEE510 pKa = 3.93TTINTHH516 pKa = 6.24GNDD519 pKa = 3.2SLIDD523 pKa = 3.53VLVIDD528 pKa = 5.61DD529 pKa = 3.99YY530 pKa = 12.06NFYY533 pKa = 10.85LSYY536 pKa = 10.62YY537 pKa = 8.61LHH539 pKa = 7.28PANFPKK545 pKa = 10.52KK546 pKa = 9.79KK547 pKa = 9.77RR548 pKa = 11.84RR549 pKa = 11.84KK550 pKa = 8.06LWYY553 pKa = 8.57MM554 pKa = 3.99
MM1 pKa = 7.25FVFLSVCVVTLIMATVNRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.87RR22 pKa = 11.84ASDD25 pKa = 2.84WDD27 pKa = 3.87LYY29 pKa = 9.78RR30 pKa = 11.84ACRR33 pKa = 11.84GGADD37 pKa = 4.17CISDD41 pKa = 3.59VVNKK45 pKa = 10.35YY46 pKa = 6.97EE47 pKa = 4.71HH48 pKa = 6.65KK49 pKa = 9.43TPADD53 pKa = 3.16KK54 pKa = 10.92VLQYY58 pKa = 11.11GGAGVFLGGLGIGTGSGALGVRR80 pKa = 11.84PGVTGPAEE88 pKa = 4.63EE89 pKa = 4.69IPLVTLGEE97 pKa = 4.29RR98 pKa = 11.84TTITDD103 pKa = 3.47TGSGRR108 pKa = 11.84TPSLGSTSFGGTSRR122 pKa = 11.84FGSGFGGRR130 pKa = 11.84IDD132 pKa = 5.37LISAGRR138 pKa = 11.84GRR140 pKa = 11.84VTTSDD145 pKa = 3.3VSVNTVDD152 pKa = 3.67SVTPSVILPEE162 pKa = 4.31TVPADD167 pKa = 3.63PGIEE171 pKa = 3.9LQVFTDD177 pKa = 3.78VEE179 pKa = 4.56DD180 pKa = 3.94SVQVTVSSSVDD191 pKa = 3.23EE192 pKa = 4.86GEE194 pKa = 4.39VAVLEE199 pKa = 4.68VPATEE204 pKa = 4.3SFGRR208 pKa = 11.84PSQVTTRR215 pKa = 11.84GGGGKK220 pKa = 8.06VHH222 pKa = 7.25HH223 pKa = 6.86SRR225 pKa = 11.84HH226 pKa = 4.92VVPAEE231 pKa = 3.67TGTILGEE238 pKa = 4.05TSEE241 pKa = 4.32TQNIFIEE248 pKa = 4.87GSGVGDD254 pKa = 3.87SYY256 pKa = 11.86SEE258 pKa = 4.63SIEE261 pKa = 3.73LWTYY265 pKa = 10.08SRR267 pKa = 11.84PRR269 pKa = 11.84YY270 pKa = 7.31STPDD274 pKa = 2.98SEE276 pKa = 4.71PPVNRR281 pKa = 11.84RR282 pKa = 11.84GINNPFSKK290 pKa = 9.95RR291 pKa = 11.84YY292 pKa = 8.0YY293 pKa = 9.57KK294 pKa = 10.3QVTVEE299 pKa = 4.61DD300 pKa = 4.29PQFLTEE306 pKa = 3.97PQKK309 pKa = 10.92LVSNVFEE316 pKa = 4.5NPAFVDD322 pKa = 4.48DD323 pKa = 4.74VDD325 pKa = 5.37DD326 pKa = 5.62SIPFLDD332 pKa = 4.11TDD334 pKa = 3.55TRR336 pKa = 11.84PYY338 pKa = 9.27NTEE341 pKa = 3.75SDD343 pKa = 3.73FLDD346 pKa = 3.22IGRR349 pKa = 11.84LGRR352 pKa = 11.84VQYY355 pKa = 9.37TVSPGEE361 pKa = 3.84GLGVSRR367 pKa = 11.84IGTRR371 pKa = 11.84FTIKK375 pKa = 9.26TRR377 pKa = 11.84SGVTIGEE384 pKa = 4.35QVHH387 pKa = 5.04YY388 pKa = 9.6RR389 pKa = 11.84YY390 pKa = 9.31PLSPITGASEE400 pKa = 5.24DD401 pKa = 3.88IEE403 pKa = 4.3LQNLVTGMSEE413 pKa = 4.38GVLDD417 pKa = 4.04SLNDD421 pKa = 3.9TVVEE425 pKa = 4.14TLNNAMLEE433 pKa = 4.32SVDD436 pKa = 5.33LDD438 pKa = 4.77DD439 pKa = 5.92ISDD442 pKa = 3.96FLSVSSSVLEE452 pKa = 4.02NEE454 pKa = 4.92LMDD457 pKa = 3.93NVEE460 pKa = 5.53DD461 pKa = 3.57ITFGQLAFRR470 pKa = 11.84DD471 pKa = 3.76EE472 pKa = 4.74GEE474 pKa = 4.57TEE476 pKa = 4.15VFTYY480 pKa = 10.37PDD482 pKa = 3.28YY483 pKa = 11.9SNTQKK488 pKa = 11.22AGVTVISEE496 pKa = 4.49DD497 pKa = 3.4NVTGTKK503 pKa = 10.28SGDD506 pKa = 3.53VVNEE510 pKa = 3.93TTINTHH516 pKa = 6.24GNDD519 pKa = 3.2SLIDD523 pKa = 3.53VLVIDD528 pKa = 5.61DD529 pKa = 3.99YY530 pKa = 12.06NFYY533 pKa = 10.85LSYY536 pKa = 10.62YY537 pKa = 8.61LHH539 pKa = 7.28PANFPKK545 pKa = 10.52KK546 pKa = 9.79KK547 pKa = 9.77RR548 pKa = 11.84RR549 pKa = 11.84KK550 pKa = 8.06LWYY553 pKa = 8.57MM554 pKa = 3.99
Molecular weight: 60.03 kDa
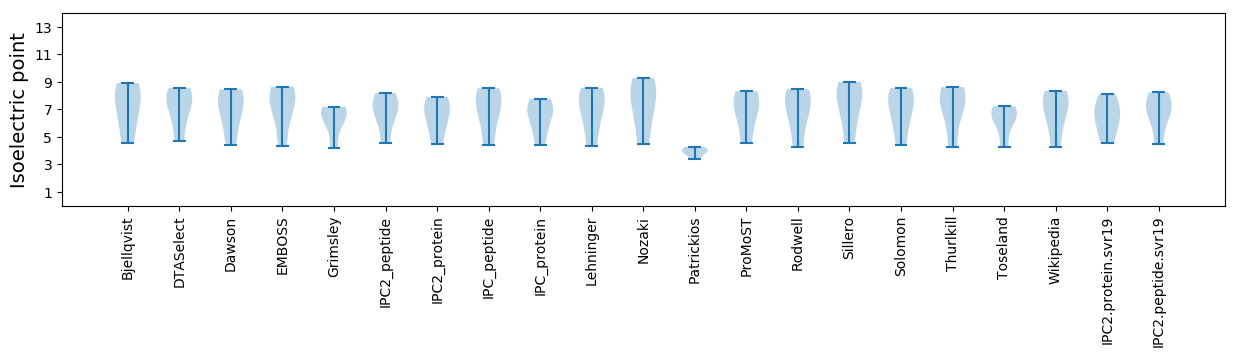
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R2Z1A9|A0A2R2Z1A9_9PAPI Regulatory protein E2 OS=Rusa timorensis papillomavirus type 2 OX=1905556 GN=E2 PE=3 SV=1
MM1 pKa = 7.0VAGPATTKK9 pKa = 10.5CLPKK13 pKa = 10.7DD14 pKa = 3.45KK15 pKa = 10.94GLGAITLTLKK25 pKa = 10.29PLPGGDD31 pKa = 3.97GVDD34 pKa = 3.62IAQKK38 pKa = 10.48APPAAQPGLKK48 pKa = 10.08NFCQLHH54 pKa = 5.8HH55 pKa = 6.86KK56 pKa = 8.49WRR58 pKa = 11.84PASNLAIQRR67 pKa = 11.84IPFADD72 pKa = 3.55LTVKK76 pKa = 10.46NYY78 pKa = 9.0YY79 pKa = 9.76VRR81 pKa = 11.84SCCPLCGQSLNYY93 pKa = 9.67CVHH96 pKa = 5.95TNKK99 pKa = 10.01TGIFSFEE106 pKa = 4.03QLLSSCFSLVCPGCAEE122 pKa = 4.12SS123 pKa = 4.31
MM1 pKa = 7.0VAGPATTKK9 pKa = 10.5CLPKK13 pKa = 10.7DD14 pKa = 3.45KK15 pKa = 10.94GLGAITLTLKK25 pKa = 10.29PLPGGDD31 pKa = 3.97GVDD34 pKa = 3.62IAQKK38 pKa = 10.48APPAAQPGLKK48 pKa = 10.08NFCQLHH54 pKa = 5.8HH55 pKa = 6.86KK56 pKa = 8.49WRR58 pKa = 11.84PASNLAIQRR67 pKa = 11.84IPFADD72 pKa = 3.55LTVKK76 pKa = 10.46NYY78 pKa = 9.0YY79 pKa = 9.76VRR81 pKa = 11.84SCCPLCGQSLNYY93 pKa = 9.67CVHH96 pKa = 5.95TNKK99 pKa = 10.01TGIFSFEE106 pKa = 4.03QLLSSCFSLVCPGCAEE122 pKa = 4.12SS123 pKa = 4.31
Molecular weight: 13.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2183 |
123 |
586 |
436.6 |
48.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.413 ± 0.954 | 2.749 ± 0.733 |
6.093 ± 0.481 | 5.772 ± 0.52 |
4.169 ± 0.603 | 7.467 ± 0.956 |
1.787 ± 0.183 | 4.26 ± 0.616 |
5.36 ± 0.783 | 8.383 ± 0.423 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.878 ± 0.415 | 4.627 ± 0.263 |
5.589 ± 0.649 | 4.031 ± 0.499 |
5.314 ± 0.303 | 7.696 ± 0.717 |
7.1 ± 0.861 | 6.505 ± 1.337 |
1.42 ± 0.29 | 3.39 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
