
Pseudoalteromonas sp. P1-26
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas; unclassified Pseudoalteromonas
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
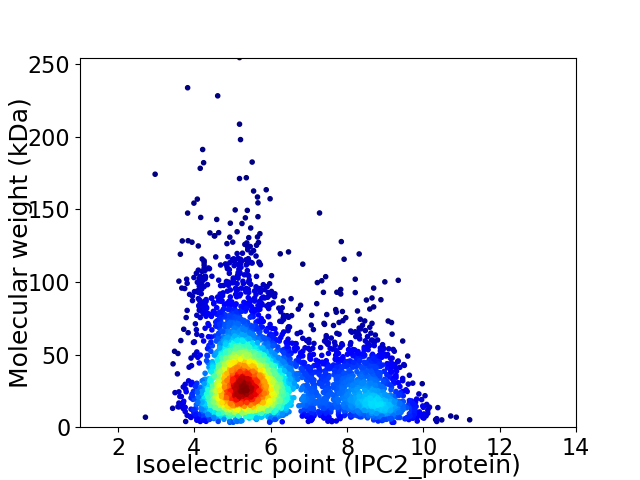
Virtual 2D-PAGE plot for 4183 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q0I066|A0A0Q0I066_9GAMM Catalase OS=Pseudoalteromonas sp. P1-26 OX=1723759 GN=katB PE=3 SV=1
MM1 pKa = 7.63QITRR5 pKa = 11.84WFFTTILVVLFSGLSYY21 pKa = 10.85SAVADD26 pKa = 3.6VVPAGTVIKK35 pKa = 9.87NQASATYY42 pKa = 7.69RR43 pKa = 11.84TCIDD47 pKa = 4.34DD48 pKa = 3.91SCSQATEE55 pKa = 4.06EE56 pKa = 5.23RR57 pKa = 11.84SITSNIVEE65 pKa = 4.28TLIQGVPAFEE75 pKa = 4.5FLTDD79 pKa = 3.33QRR81 pKa = 11.84LPAISDD87 pKa = 3.49RR88 pKa = 11.84PVYY91 pKa = 9.63FAHH94 pKa = 6.93TISNIGNVADD104 pKa = 5.2RR105 pKa = 11.84YY106 pKa = 8.86QICVSAVDD114 pKa = 3.79AQVSEE119 pKa = 3.85WRR121 pKa = 11.84VYY123 pKa = 11.55ADD125 pKa = 3.2TDD127 pKa = 3.93LDD129 pKa = 3.8GRR131 pKa = 11.84PDD133 pKa = 3.76PGQILLSSDD142 pKa = 4.21DD143 pKa = 4.49GSACLPNLTNSIDD156 pKa = 3.78PSEE159 pKa = 4.12TWGLVIEE166 pKa = 4.89VIANGAAGSVLLSAIEE182 pKa = 4.29LEE184 pKa = 4.52ATSSDD189 pKa = 4.67DD190 pKa = 3.29NTLNQTVVDD199 pKa = 3.91SVEE202 pKa = 4.7FIEE205 pKa = 5.4GPLIEE210 pKa = 4.75VLKK213 pKa = 10.83SMPVRR218 pKa = 11.84VGGSPSGPYY227 pKa = 8.31TVRR230 pKa = 11.84LAYY233 pKa = 10.57RR234 pKa = 11.84NVSNFTASSVVLEE247 pKa = 5.54DD248 pKa = 3.6ILPTTFFDD256 pKa = 5.06GQGQPQTGGFEE267 pKa = 4.27YY268 pKa = 11.06VPGSGRR274 pKa = 11.84WSQTGNTALTDD285 pKa = 4.06DD286 pKa = 4.82DD287 pKa = 5.65DD288 pKa = 4.61GVEE291 pKa = 4.35GGGTEE296 pKa = 3.91PASYY300 pKa = 10.14CAYY303 pKa = 10.49DD304 pKa = 3.61GTAANVDD311 pKa = 3.52CQDD314 pKa = 3.02RR315 pKa = 11.84VRR317 pKa = 11.84FEE319 pKa = 4.68LATLPPGGEE328 pKa = 4.4GYY330 pKa = 9.92IEE332 pKa = 4.31FAVQVVGGISTGGRR346 pKa = 11.84ILNIAGYY353 pKa = 10.2YY354 pKa = 8.99YY355 pKa = 10.92NNEE358 pKa = 4.39DD359 pKa = 3.33NTVVYY364 pKa = 9.46AQSGSGPVGGAITPFDD380 pKa = 4.04TNVVPFQIKK389 pKa = 10.16SSATAPSVVVNDD401 pKa = 3.62SSTDD405 pKa = 3.25INIGVDD411 pKa = 3.53DD412 pKa = 4.84SLDD415 pKa = 3.36SGNRR419 pKa = 11.84VEE421 pKa = 5.57IAAAGQGAVIRR432 pKa = 11.84FTNYY436 pKa = 8.14IWNTGDD442 pKa = 3.91GVDD445 pKa = 3.84TFDD448 pKa = 5.95LNVDD452 pKa = 3.59RR453 pKa = 11.84ALNRR457 pKa = 11.84VGASLANPFPDD468 pKa = 3.34NAIVRR473 pKa = 11.84VLQADD478 pKa = 4.28GATPLLDD485 pKa = 3.41TSNSGRR491 pKa = 11.84VDD493 pKa = 3.41TGPIPLPNFDD503 pKa = 4.25TGACSGRR510 pKa = 11.84FVTSADD516 pKa = 3.25NTRR519 pKa = 11.84CGLPVVVEE527 pKa = 4.24VEE529 pKa = 4.33LPADD533 pKa = 3.67AVGGPYY539 pKa = 10.61EE540 pKa = 4.3LTLAATSNVDD550 pKa = 3.41TTVTNAVSNILLSVVPATVDD570 pKa = 2.98ITNNLPLSAGSAPGAGVGPEE590 pKa = 3.98PTAVTTISLMPGNTGTFTLYY610 pKa = 11.06VNNTGDD616 pKa = 4.34RR617 pKa = 11.84QDD619 pKa = 3.55QYY621 pKa = 11.98QLAASTTNFAAGTLYY636 pKa = 10.22PGWLVSFYY644 pKa = 10.65RR645 pKa = 11.84DD646 pKa = 3.55GGAGDD651 pKa = 4.13CTSLGNAIDD660 pKa = 3.72VTDD663 pKa = 5.48IIPAGSAQLVCAHH676 pKa = 5.94VQTAANAEE684 pKa = 3.88VDD686 pKa = 3.95VYY688 pKa = 11.6DD689 pKa = 4.22LYY691 pKa = 11.42FRR693 pKa = 11.84ASSLLSAAQDD703 pKa = 3.18IKK705 pKa = 11.32LDD707 pKa = 3.51AVEE710 pKa = 4.86ILGAPDD716 pKa = 3.46ISFSPDD722 pKa = 2.58QFNQVVPGSSVSYY735 pKa = 8.02THH737 pKa = 6.58QIKK740 pKa = 8.18NTGTVPLSSIQLTATPQAEE759 pKa = 4.83DD760 pKa = 3.71DD761 pKa = 4.2NGWAVLVYY769 pKa = 10.29EE770 pKa = 4.92DD771 pKa = 5.31DD772 pKa = 4.6GNGVWGPEE780 pKa = 4.24DD781 pKa = 3.37EE782 pKa = 4.74TIPLGGVLGTDD793 pKa = 3.23NGDD796 pKa = 3.8EE797 pKa = 3.87ILLPGEE803 pKa = 4.66SITVHH808 pKa = 5.0VRR810 pKa = 11.84VFAPASAVLGDD821 pKa = 4.1INVKK825 pKa = 9.72TITLTTTVGSDD836 pKa = 2.93TLEE839 pKa = 4.25RR840 pKa = 11.84SVTDD844 pKa = 3.26STSTSQGNTLIFKK857 pKa = 9.49EE858 pKa = 3.89QALDD862 pKa = 3.96SNCDD866 pKa = 3.51GVPDD870 pKa = 4.98GPLACTGDD878 pKa = 3.92SCFSTSVFPVHH889 pKa = 7.14PGQCVLYY896 pKa = 10.28RR897 pKa = 11.84LTARR901 pKa = 11.84NQSAEE906 pKa = 3.88PVFNVLINDD915 pKa = 3.76RR916 pKa = 11.84TQAYY920 pKa = 5.48TTYY923 pKa = 10.37FQTALNCSSPSGSCNADD940 pKa = 2.73VTTPNDD946 pKa = 3.59GGVGDD951 pKa = 4.11IQADD955 pKa = 3.43IGVLQAGEE963 pKa = 3.79QAILIFGVRR972 pKa = 11.84VRR974 pKa = 4.25
MM1 pKa = 7.63QITRR5 pKa = 11.84WFFTTILVVLFSGLSYY21 pKa = 10.85SAVADD26 pKa = 3.6VVPAGTVIKK35 pKa = 9.87NQASATYY42 pKa = 7.69RR43 pKa = 11.84TCIDD47 pKa = 4.34DD48 pKa = 3.91SCSQATEE55 pKa = 4.06EE56 pKa = 5.23RR57 pKa = 11.84SITSNIVEE65 pKa = 4.28TLIQGVPAFEE75 pKa = 4.5FLTDD79 pKa = 3.33QRR81 pKa = 11.84LPAISDD87 pKa = 3.49RR88 pKa = 11.84PVYY91 pKa = 9.63FAHH94 pKa = 6.93TISNIGNVADD104 pKa = 5.2RR105 pKa = 11.84YY106 pKa = 8.86QICVSAVDD114 pKa = 3.79AQVSEE119 pKa = 3.85WRR121 pKa = 11.84VYY123 pKa = 11.55ADD125 pKa = 3.2TDD127 pKa = 3.93LDD129 pKa = 3.8GRR131 pKa = 11.84PDD133 pKa = 3.76PGQILLSSDD142 pKa = 4.21DD143 pKa = 4.49GSACLPNLTNSIDD156 pKa = 3.78PSEE159 pKa = 4.12TWGLVIEE166 pKa = 4.89VIANGAAGSVLLSAIEE182 pKa = 4.29LEE184 pKa = 4.52ATSSDD189 pKa = 4.67DD190 pKa = 3.29NTLNQTVVDD199 pKa = 3.91SVEE202 pKa = 4.7FIEE205 pKa = 5.4GPLIEE210 pKa = 4.75VLKK213 pKa = 10.83SMPVRR218 pKa = 11.84VGGSPSGPYY227 pKa = 8.31TVRR230 pKa = 11.84LAYY233 pKa = 10.57RR234 pKa = 11.84NVSNFTASSVVLEE247 pKa = 5.54DD248 pKa = 3.6ILPTTFFDD256 pKa = 5.06GQGQPQTGGFEE267 pKa = 4.27YY268 pKa = 11.06VPGSGRR274 pKa = 11.84WSQTGNTALTDD285 pKa = 4.06DD286 pKa = 4.82DD287 pKa = 5.65DD288 pKa = 4.61GVEE291 pKa = 4.35GGGTEE296 pKa = 3.91PASYY300 pKa = 10.14CAYY303 pKa = 10.49DD304 pKa = 3.61GTAANVDD311 pKa = 3.52CQDD314 pKa = 3.02RR315 pKa = 11.84VRR317 pKa = 11.84FEE319 pKa = 4.68LATLPPGGEE328 pKa = 4.4GYY330 pKa = 9.92IEE332 pKa = 4.31FAVQVVGGISTGGRR346 pKa = 11.84ILNIAGYY353 pKa = 10.2YY354 pKa = 8.99YY355 pKa = 10.92NNEE358 pKa = 4.39DD359 pKa = 3.33NTVVYY364 pKa = 9.46AQSGSGPVGGAITPFDD380 pKa = 4.04TNVVPFQIKK389 pKa = 10.16SSATAPSVVVNDD401 pKa = 3.62SSTDD405 pKa = 3.25INIGVDD411 pKa = 3.53DD412 pKa = 4.84SLDD415 pKa = 3.36SGNRR419 pKa = 11.84VEE421 pKa = 5.57IAAAGQGAVIRR432 pKa = 11.84FTNYY436 pKa = 8.14IWNTGDD442 pKa = 3.91GVDD445 pKa = 3.84TFDD448 pKa = 5.95LNVDD452 pKa = 3.59RR453 pKa = 11.84ALNRR457 pKa = 11.84VGASLANPFPDD468 pKa = 3.34NAIVRR473 pKa = 11.84VLQADD478 pKa = 4.28GATPLLDD485 pKa = 3.41TSNSGRR491 pKa = 11.84VDD493 pKa = 3.41TGPIPLPNFDD503 pKa = 4.25TGACSGRR510 pKa = 11.84FVTSADD516 pKa = 3.25NTRR519 pKa = 11.84CGLPVVVEE527 pKa = 4.24VEE529 pKa = 4.33LPADD533 pKa = 3.67AVGGPYY539 pKa = 10.61EE540 pKa = 4.3LTLAATSNVDD550 pKa = 3.41TTVTNAVSNILLSVVPATVDD570 pKa = 2.98ITNNLPLSAGSAPGAGVGPEE590 pKa = 3.98PTAVTTISLMPGNTGTFTLYY610 pKa = 11.06VNNTGDD616 pKa = 4.34RR617 pKa = 11.84QDD619 pKa = 3.55QYY621 pKa = 11.98QLAASTTNFAAGTLYY636 pKa = 10.22PGWLVSFYY644 pKa = 10.65RR645 pKa = 11.84DD646 pKa = 3.55GGAGDD651 pKa = 4.13CTSLGNAIDD660 pKa = 3.72VTDD663 pKa = 5.48IIPAGSAQLVCAHH676 pKa = 5.94VQTAANAEE684 pKa = 3.88VDD686 pKa = 3.95VYY688 pKa = 11.6DD689 pKa = 4.22LYY691 pKa = 11.42FRR693 pKa = 11.84ASSLLSAAQDD703 pKa = 3.18IKK705 pKa = 11.32LDD707 pKa = 3.51AVEE710 pKa = 4.86ILGAPDD716 pKa = 3.46ISFSPDD722 pKa = 2.58QFNQVVPGSSVSYY735 pKa = 8.02THH737 pKa = 6.58QIKK740 pKa = 8.18NTGTVPLSSIQLTATPQAEE759 pKa = 4.83DD760 pKa = 3.71DD761 pKa = 4.2NGWAVLVYY769 pKa = 10.29EE770 pKa = 4.92DD771 pKa = 5.31DD772 pKa = 4.6GNGVWGPEE780 pKa = 4.24DD781 pKa = 3.37EE782 pKa = 4.74TIPLGGVLGTDD793 pKa = 3.23NGDD796 pKa = 3.8EE797 pKa = 3.87ILLPGEE803 pKa = 4.66SITVHH808 pKa = 5.0VRR810 pKa = 11.84VFAPASAVLGDD821 pKa = 4.1INVKK825 pKa = 9.72TITLTTTVGSDD836 pKa = 2.93TLEE839 pKa = 4.25RR840 pKa = 11.84SVTDD844 pKa = 3.26STSTSQGNTLIFKK857 pKa = 9.49EE858 pKa = 3.89QALDD862 pKa = 3.96SNCDD866 pKa = 3.51GVPDD870 pKa = 4.98GPLACTGDD878 pKa = 3.92SCFSTSVFPVHH889 pKa = 7.14PGQCVLYY896 pKa = 10.28RR897 pKa = 11.84LTARR901 pKa = 11.84NQSAEE906 pKa = 3.88PVFNVLINDD915 pKa = 3.76RR916 pKa = 11.84TQAYY920 pKa = 5.48TTYY923 pKa = 10.37FQTALNCSSPSGSCNADD940 pKa = 2.73VTTPNDD946 pKa = 3.59GGVGDD951 pKa = 4.11IQADD955 pKa = 3.43IGVLQAGEE963 pKa = 3.79QAILIFGVRR972 pKa = 11.84VRR974 pKa = 4.25
Molecular weight: 101.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q0J7Z7|A0A0Q0J7Z7_9GAMM Ribose-phosphate pyrophosphokinase OS=Pseudoalteromonas sp. P1-26 OX=1723759 GN=prs_1 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1392368 |
29 |
2234 |
332.9 |
37.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.853 ± 0.043 | 0.989 ± 0.013 |
5.741 ± 0.04 | 6.213 ± 0.036 |
4.383 ± 0.022 | 6.401 ± 0.039 |
2.205 ± 0.02 | 6.296 ± 0.035 |
5.701 ± 0.035 | 10.414 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.362 ± 0.018 | 4.656 ± 0.026 |
3.638 ± 0.021 | 4.792 ± 0.035 |
3.978 ± 0.029 | 6.785 ± 0.035 |
5.401 ± 0.029 | 6.757 ± 0.028 |
1.153 ± 0.014 | 3.283 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
