
Massilia namucuonensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Massilia
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
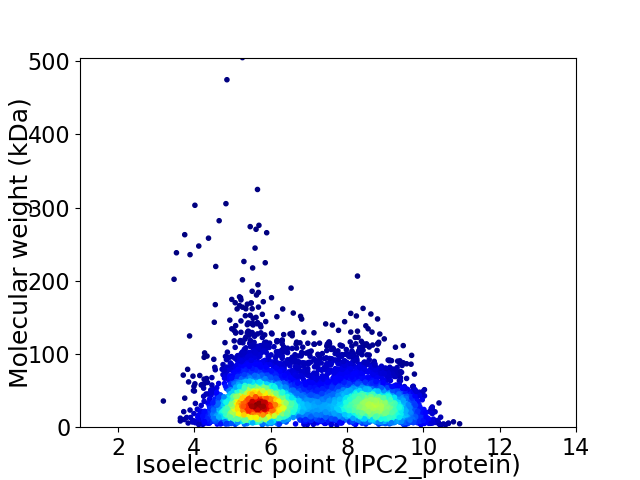
Virtual 2D-PAGE plot for 7034 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
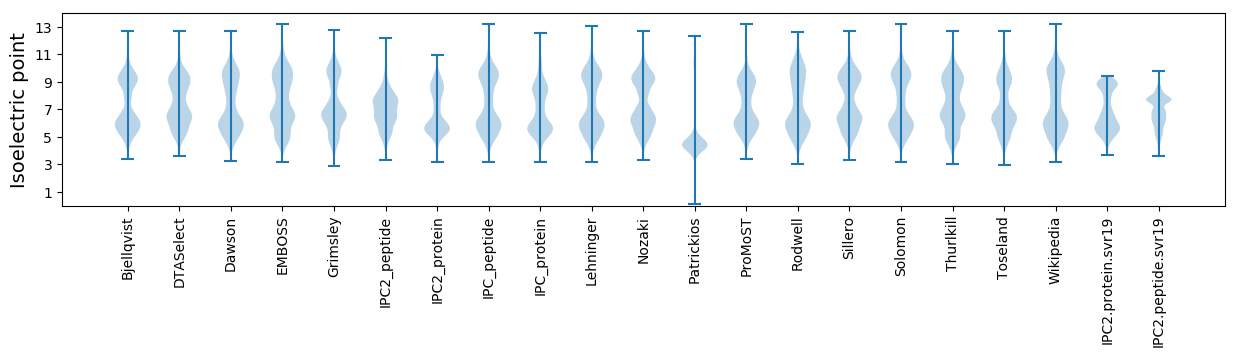
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I7FS64|A0A1I7FS64_9BURK DUF4214 domain-containing protein OS=Massilia namucuonensis OX=1035707 GN=SAMN05216552_1002247 PE=4 SV=1
MM1 pKa = 7.43TAFKK5 pKa = 9.91WVAGEE10 pKa = 4.27TKK12 pKa = 10.3SIQYY16 pKa = 10.41AAGDD20 pKa = 3.82TLEE23 pKa = 4.59LSGIAASQLEE33 pKa = 4.38LSVADD38 pKa = 4.23GRR40 pKa = 11.84LLLAAGALGTLTLTPVAGGALTAASLALTFPDD72 pKa = 3.32GGRR75 pKa = 11.84YY76 pKa = 8.78IPAGPDD82 pKa = 2.96ALTEE86 pKa = 4.5GGDD89 pKa = 3.56GGDD92 pKa = 3.79YY93 pKa = 11.18LLGDD97 pKa = 3.82DD98 pKa = 4.68QGNFVDD104 pKa = 5.77AGLGDD109 pKa = 3.89DD110 pKa = 3.78RR111 pKa = 11.84VEE113 pKa = 4.4GGGDD117 pKa = 3.46GDD119 pKa = 4.18YY120 pKa = 11.23LIGGAGDD127 pKa = 4.28DD128 pKa = 4.27LLLGGGGDD136 pKa = 4.25DD137 pKa = 5.02LLAGGDD143 pKa = 4.11GRR145 pKa = 11.84DD146 pKa = 3.72TLDD149 pKa = 4.12GGDD152 pKa = 3.96GANLLLGQKK161 pKa = 10.55GDD163 pKa = 3.68DD164 pKa = 3.6TYY166 pKa = 11.64LVHH169 pKa = 6.96SQDD172 pKa = 5.5DD173 pKa = 4.4YY174 pKa = 11.64ISDD177 pKa = 3.62SGGVDD182 pKa = 3.03GGVINADD189 pKa = 3.04WYY191 pKa = 9.0KK192 pKa = 10.04TADD195 pKa = 3.66EE196 pKa = 4.47VEE198 pKa = 4.02NWTWAPGVQRR208 pKa = 11.84LPYY211 pKa = 9.53WVDD214 pKa = 3.21ALTADD219 pKa = 3.83YY220 pKa = 10.85APYY223 pKa = 10.58VAMLVRR229 pKa = 11.84TAGRR233 pKa = 11.84SVHH236 pKa = 6.07YY237 pKa = 10.09SFPTTPADD245 pKa = 4.37FFTDD249 pKa = 3.2DD250 pKa = 4.36DD251 pKa = 4.55RR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.75FLPFTEE260 pKa = 4.72DD261 pKa = 2.84QIAYY265 pKa = 6.38TRR267 pKa = 11.84KK268 pKa = 8.23ALAYY272 pKa = 9.53IEE274 pKa = 4.32TVLNIKK280 pKa = 10.35FEE282 pKa = 4.38EE283 pKa = 4.44TDD285 pKa = 3.94SPDD288 pKa = 3.29QPLSIVFGNNRR299 pKa = 11.84QEE301 pKa = 4.85DD302 pKa = 3.81SAAYY306 pKa = 10.16ANSLHH311 pKa = 7.49DD312 pKa = 4.99DD313 pKa = 4.05FGSPLMLSEE322 pKa = 5.42DD323 pKa = 4.38FLVMHH328 pKa = 7.19PSRR331 pKa = 11.84DD332 pKa = 3.94DD333 pKa = 3.88GDD335 pKa = 3.96SLYY338 pKa = 11.3SLLMHH343 pKa = 6.8EE344 pKa = 5.41LGHH347 pKa = 6.39SLGLKK352 pKa = 10.14HH353 pKa = 6.65PFAAPDD359 pKa = 3.46AYY361 pKa = 10.4GGEE364 pKa = 4.35IGAGPYY370 pKa = 10.01LGSGEE375 pKa = 4.66DD376 pKa = 3.46QTDD379 pKa = 3.6ATVMSYY385 pKa = 10.0TEE387 pKa = 4.64GEE389 pKa = 3.91LAQYY393 pKa = 11.3DD394 pKa = 4.12RR395 pKa = 11.84FSPLDD400 pKa = 3.44IAALQYY406 pKa = 10.98LYY408 pKa = 11.04GVAPGATGARR418 pKa = 11.84THH420 pKa = 6.15VLVATAANMIWVGAGLNIIDD440 pKa = 4.59GAHH443 pKa = 6.0LTADD447 pKa = 3.5LTLNLEE453 pKa = 4.52PGHH456 pKa = 6.59WSHH459 pKa = 7.55IGAKK463 pKa = 10.07ADD465 pKa = 3.87TITAAGQITINFGSVIVGAKK485 pKa = 10.01GGAGNDD491 pKa = 3.47ALTGNAISNEE501 pKa = 3.97LFGGAGNDD509 pKa = 3.83TLSGGASDD517 pKa = 5.68DD518 pKa = 3.85YY519 pKa = 11.74LFGGAGNDD527 pKa = 3.5ILSGGADD534 pKa = 3.64PDD536 pKa = 4.19YY537 pKa = 11.34LVGDD541 pKa = 4.98AGNDD545 pKa = 3.69MLSGGADD552 pKa = 3.45PDD554 pKa = 4.08YY555 pKa = 11.46LFGEE559 pKa = 4.74AGNDD563 pKa = 3.7TLSGGAEE570 pKa = 3.81ADD572 pKa = 3.57QLFGGAGVDD581 pKa = 3.36TAVFTGARR589 pKa = 11.84RR590 pKa = 11.84AYY592 pKa = 10.34DD593 pKa = 3.55LAWQNEE599 pKa = 4.07ALTVSALAGMDD610 pKa = 3.57GTDD613 pKa = 3.15TLNDD617 pKa = 3.34VEE619 pKa = 4.72RR620 pKa = 11.84LRR622 pKa = 11.84FADD625 pKa = 4.52GMLALDD631 pKa = 3.98VGGDD635 pKa = 3.69GAGGQVYY642 pKa = 10.17RR643 pKa = 11.84LYY645 pKa = 10.66QAAFDD650 pKa = 4.52RR651 pKa = 11.84KK652 pKa = 9.31PDD654 pKa = 3.76AAGVGYY660 pKa = 9.55WLAQADD666 pKa = 4.04QGAGMAGIAEE676 pKa = 4.19SFLRR680 pKa = 11.84SEE682 pKa = 4.24EE683 pKa = 3.85FARR686 pKa = 11.84LYY688 pKa = 11.09GGANPTDD695 pKa = 3.24AAFVAKK701 pKa = 10.39LYY703 pKa = 11.1SNVLHH708 pKa = 6.98RR709 pKa = 11.84AYY711 pKa = 10.58DD712 pKa = 3.55QTGYY716 pKa = 11.1DD717 pKa = 3.32YY718 pKa = 7.49WTWTLAAGASRR729 pKa = 11.84AEE731 pKa = 4.12VLLSFAAGEE740 pKa = 4.01EE741 pKa = 4.41NYY743 pKa = 10.2QQVAAIIANGYY754 pKa = 9.45EE755 pKa = 4.02YY756 pKa = 11.36AHH758 pKa = 6.52YY759 pKa = 10.67GG760 pKa = 3.38
MM1 pKa = 7.43TAFKK5 pKa = 9.91WVAGEE10 pKa = 4.27TKK12 pKa = 10.3SIQYY16 pKa = 10.41AAGDD20 pKa = 3.82TLEE23 pKa = 4.59LSGIAASQLEE33 pKa = 4.38LSVADD38 pKa = 4.23GRR40 pKa = 11.84LLLAAGALGTLTLTPVAGGALTAASLALTFPDD72 pKa = 3.32GGRR75 pKa = 11.84YY76 pKa = 8.78IPAGPDD82 pKa = 2.96ALTEE86 pKa = 4.5GGDD89 pKa = 3.56GGDD92 pKa = 3.79YY93 pKa = 11.18LLGDD97 pKa = 3.82DD98 pKa = 4.68QGNFVDD104 pKa = 5.77AGLGDD109 pKa = 3.89DD110 pKa = 3.78RR111 pKa = 11.84VEE113 pKa = 4.4GGGDD117 pKa = 3.46GDD119 pKa = 4.18YY120 pKa = 11.23LIGGAGDD127 pKa = 4.28DD128 pKa = 4.27LLLGGGGDD136 pKa = 4.25DD137 pKa = 5.02LLAGGDD143 pKa = 4.11GRR145 pKa = 11.84DD146 pKa = 3.72TLDD149 pKa = 4.12GGDD152 pKa = 3.96GANLLLGQKK161 pKa = 10.55GDD163 pKa = 3.68DD164 pKa = 3.6TYY166 pKa = 11.64LVHH169 pKa = 6.96SQDD172 pKa = 5.5DD173 pKa = 4.4YY174 pKa = 11.64ISDD177 pKa = 3.62SGGVDD182 pKa = 3.03GGVINADD189 pKa = 3.04WYY191 pKa = 9.0KK192 pKa = 10.04TADD195 pKa = 3.66EE196 pKa = 4.47VEE198 pKa = 4.02NWTWAPGVQRR208 pKa = 11.84LPYY211 pKa = 9.53WVDD214 pKa = 3.21ALTADD219 pKa = 3.83YY220 pKa = 10.85APYY223 pKa = 10.58VAMLVRR229 pKa = 11.84TAGRR233 pKa = 11.84SVHH236 pKa = 6.07YY237 pKa = 10.09SFPTTPADD245 pKa = 4.37FFTDD249 pKa = 3.2DD250 pKa = 4.36DD251 pKa = 4.55RR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.75FLPFTEE260 pKa = 4.72DD261 pKa = 2.84QIAYY265 pKa = 6.38TRR267 pKa = 11.84KK268 pKa = 8.23ALAYY272 pKa = 9.53IEE274 pKa = 4.32TVLNIKK280 pKa = 10.35FEE282 pKa = 4.38EE283 pKa = 4.44TDD285 pKa = 3.94SPDD288 pKa = 3.29QPLSIVFGNNRR299 pKa = 11.84QEE301 pKa = 4.85DD302 pKa = 3.81SAAYY306 pKa = 10.16ANSLHH311 pKa = 7.49DD312 pKa = 4.99DD313 pKa = 4.05FGSPLMLSEE322 pKa = 5.42DD323 pKa = 4.38FLVMHH328 pKa = 7.19PSRR331 pKa = 11.84DD332 pKa = 3.94DD333 pKa = 3.88GDD335 pKa = 3.96SLYY338 pKa = 11.3SLLMHH343 pKa = 6.8EE344 pKa = 5.41LGHH347 pKa = 6.39SLGLKK352 pKa = 10.14HH353 pKa = 6.65PFAAPDD359 pKa = 3.46AYY361 pKa = 10.4GGEE364 pKa = 4.35IGAGPYY370 pKa = 10.01LGSGEE375 pKa = 4.66DD376 pKa = 3.46QTDD379 pKa = 3.6ATVMSYY385 pKa = 10.0TEE387 pKa = 4.64GEE389 pKa = 3.91LAQYY393 pKa = 11.3DD394 pKa = 4.12RR395 pKa = 11.84FSPLDD400 pKa = 3.44IAALQYY406 pKa = 10.98LYY408 pKa = 11.04GVAPGATGARR418 pKa = 11.84THH420 pKa = 6.15VLVATAANMIWVGAGLNIIDD440 pKa = 4.59GAHH443 pKa = 6.0LTADD447 pKa = 3.5LTLNLEE453 pKa = 4.52PGHH456 pKa = 6.59WSHH459 pKa = 7.55IGAKK463 pKa = 10.07ADD465 pKa = 3.87TITAAGQITINFGSVIVGAKK485 pKa = 10.01GGAGNDD491 pKa = 3.47ALTGNAISNEE501 pKa = 3.97LFGGAGNDD509 pKa = 3.83TLSGGASDD517 pKa = 5.68DD518 pKa = 3.85YY519 pKa = 11.74LFGGAGNDD527 pKa = 3.5ILSGGADD534 pKa = 3.64PDD536 pKa = 4.19YY537 pKa = 11.34LVGDD541 pKa = 4.98AGNDD545 pKa = 3.69MLSGGADD552 pKa = 3.45PDD554 pKa = 4.08YY555 pKa = 11.46LFGEE559 pKa = 4.74AGNDD563 pKa = 3.7TLSGGAEE570 pKa = 3.81ADD572 pKa = 3.57QLFGGAGVDD581 pKa = 3.36TAVFTGARR589 pKa = 11.84RR590 pKa = 11.84AYY592 pKa = 10.34DD593 pKa = 3.55LAWQNEE599 pKa = 4.07ALTVSALAGMDD610 pKa = 3.57GTDD613 pKa = 3.15TLNDD617 pKa = 3.34VEE619 pKa = 4.72RR620 pKa = 11.84LRR622 pKa = 11.84FADD625 pKa = 4.52GMLALDD631 pKa = 3.98VGGDD635 pKa = 3.69GAGGQVYY642 pKa = 10.17RR643 pKa = 11.84LYY645 pKa = 10.66QAAFDD650 pKa = 4.52RR651 pKa = 11.84KK652 pKa = 9.31PDD654 pKa = 3.76AAGVGYY660 pKa = 9.55WLAQADD666 pKa = 4.04QGAGMAGIAEE676 pKa = 4.19SFLRR680 pKa = 11.84SEE682 pKa = 4.24EE683 pKa = 3.85FARR686 pKa = 11.84LYY688 pKa = 11.09GGANPTDD695 pKa = 3.24AAFVAKK701 pKa = 10.39LYY703 pKa = 11.1SNVLHH708 pKa = 6.98RR709 pKa = 11.84AYY711 pKa = 10.58DD712 pKa = 3.55QTGYY716 pKa = 11.1DD717 pKa = 3.32YY718 pKa = 7.49WTWTLAAGASRR729 pKa = 11.84AEE731 pKa = 4.12VLLSFAAGEE740 pKa = 4.01EE741 pKa = 4.41NYY743 pKa = 10.2QQVAAIIANGYY754 pKa = 9.45EE755 pKa = 4.02YY756 pKa = 11.36AHH758 pKa = 6.52YY759 pKa = 10.67GG760 pKa = 3.38
Molecular weight: 79.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I7M7E5|A0A1I7M7E5_9BURK Uncharacterized protein OS=Massilia namucuonensis OX=1035707 GN=SAMN05216552_10725 PE=4 SV=1
II1 pKa = 7.27RR2 pKa = 11.84VKK4 pKa = 10.56LLKK7 pKa = 10.22IGAAIVRR14 pKa = 11.84NTRR17 pKa = 11.84RR18 pKa = 11.84VRR20 pKa = 11.84VLLASQHH27 pKa = 5.35PLKK30 pKa = 10.79HH31 pKa = 5.95VFISAARR38 pKa = 11.84AMAPP42 pKa = 3.32
II1 pKa = 7.27RR2 pKa = 11.84VKK4 pKa = 10.56LLKK7 pKa = 10.22IGAAIVRR14 pKa = 11.84NTRR17 pKa = 11.84RR18 pKa = 11.84VRR20 pKa = 11.84VLLASQHH27 pKa = 5.35PLKK30 pKa = 10.79HH31 pKa = 5.95VFISAARR38 pKa = 11.84AMAPP42 pKa = 3.32
Molecular weight: 4.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2567245 |
27 |
4738 |
365.0 |
39.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.327 ± 0.059 | 0.857 ± 0.009 |
5.392 ± 0.02 | 5.201 ± 0.029 |
3.416 ± 0.019 | 8.691 ± 0.038 |
2.167 ± 0.014 | 4.327 ± 0.023 |
3.47 ± 0.028 | 10.329 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.517 ± 0.015 | 3.021 ± 0.021 |
5.088 ± 0.026 | 3.733 ± 0.023 |
6.746 ± 0.037 | 5.398 ± 0.025 |
5.189 ± 0.037 | 7.173 ± 0.024 |
1.364 ± 0.012 | 2.593 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
