
Beihai weivirus-like virus 13
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
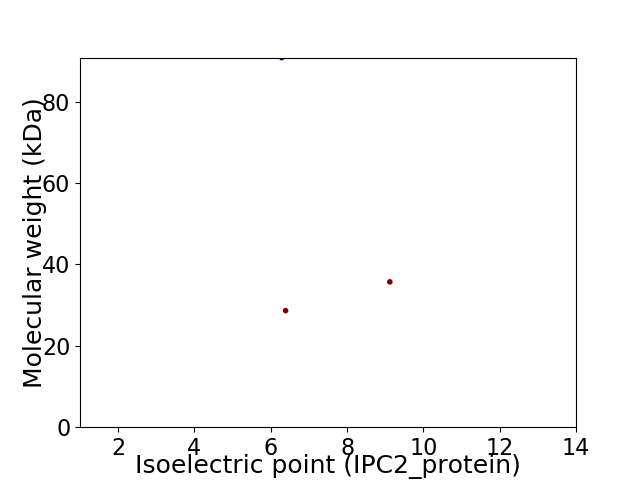
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
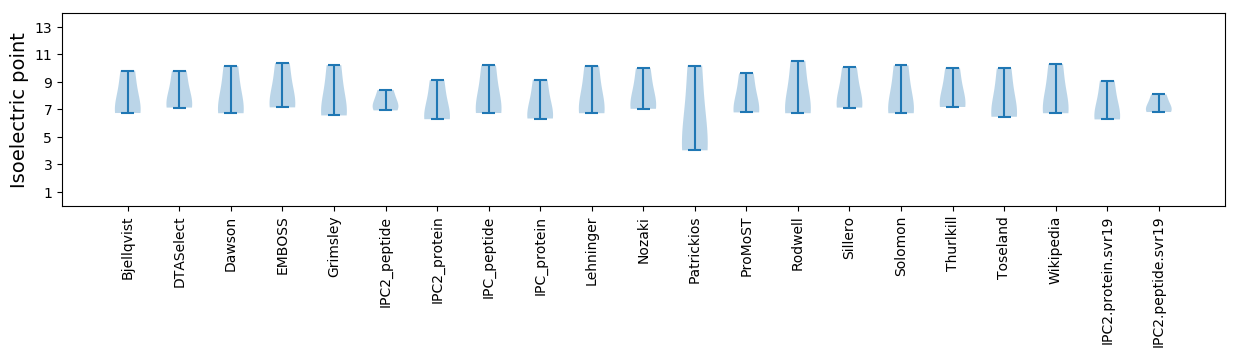
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL24|A0A1L3KL24_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 13 OX=1922742 PE=4 SV=1
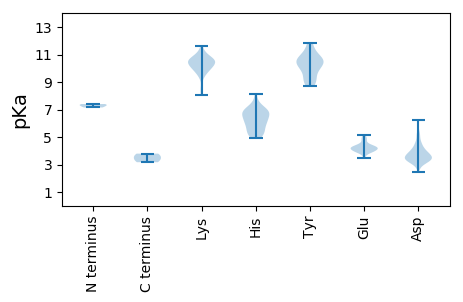
MM1 pKa = 7.4AVVSFRR7 pKa = 11.84PPSRR11 pKa = 11.84RR12 pKa = 11.84NDD14 pKa = 3.17LCYY17 pKa = 10.43EE18 pKa = 4.09CHH20 pKa = 6.38SWMLQHH26 pKa = 6.55LTYY29 pKa = 10.58EE30 pKa = 4.45HH31 pKa = 6.92ASGRR35 pKa = 11.84ADD37 pKa = 3.2VSDD40 pKa = 3.43TFLYY44 pKa = 10.54EE45 pKa = 5.1LEE47 pKa = 4.33LQVARR52 pKa = 11.84CGRR55 pKa = 11.84HH56 pKa = 5.52VEE58 pKa = 4.31TCAQRR63 pKa = 11.84IRR65 pKa = 11.84DD66 pKa = 3.41RR67 pKa = 11.84LGRR70 pKa = 11.84IKK72 pKa = 10.5HH73 pKa = 5.49ALCGQFHH80 pKa = 6.48GVGVHH85 pKa = 5.93HH86 pKa = 7.26VYY88 pKa = 9.12LTRR91 pKa = 11.84GEE93 pKa = 3.96VDD95 pKa = 2.47IMRR98 pKa = 11.84RR99 pKa = 11.84SGGAALYY106 pKa = 10.55SGAPTTTADD115 pKa = 3.86SNLHH119 pKa = 5.44FATYY123 pKa = 10.7RR124 pKa = 11.84LDD126 pKa = 3.46KK127 pKa = 10.66KK128 pKa = 9.56EE129 pKa = 3.87RR130 pKa = 11.84RR131 pKa = 11.84GDD133 pKa = 4.12PISCATIDD141 pKa = 3.6QLFGYY146 pKa = 9.73LAATIQQGRR155 pKa = 11.84VANRR159 pKa = 11.84HH160 pKa = 5.46CCCLHH165 pKa = 5.34TAGLGLRR172 pKa = 11.84NLRR175 pKa = 11.84PVKK178 pKa = 10.26RR179 pKa = 11.84MPEE182 pKa = 4.11DD183 pKa = 3.41EE184 pKa = 4.7PEE186 pKa = 4.26PPKK189 pKa = 10.96SDD191 pKa = 3.43DD192 pKa = 3.73GKK194 pKa = 10.42DD195 pKa = 3.57GKK197 pKa = 11.18VEE199 pKa = 4.29GPPEE203 pKa = 4.82DD204 pKa = 5.43DD205 pKa = 4.36PPQVDD210 pKa = 3.79CHH212 pKa = 6.11TCHH215 pKa = 6.82SGLAGIQLIHH225 pKa = 7.18DD226 pKa = 4.1VNEE229 pKa = 4.5GPEE232 pKa = 4.32TIPLAVRR239 pKa = 11.84YY240 pKa = 9.05LPQIEE245 pKa = 4.96EE246 pKa = 3.94RR247 pKa = 11.84LFWLNSQKK255 pKa = 10.66NVEE258 pKa = 4.1RR259 pKa = 11.84AIKK262 pKa = 9.85EE263 pKa = 4.18RR264 pKa = 11.84IEE266 pKa = 4.18EE267 pKa = 3.92PHH269 pKa = 6.83VPVTLSQEE277 pKa = 4.16EE278 pKa = 4.23KK279 pKa = 10.65DD280 pKa = 3.82DD281 pKa = 3.98LRR283 pKa = 11.84AVTQVMIDD291 pKa = 3.3QLKK294 pKa = 9.97ADD296 pKa = 4.47KK297 pKa = 11.16GLIDD301 pKa = 6.26KK302 pKa = 10.53IIQDD306 pKa = 3.55KK307 pKa = 10.98LGIYY311 pKa = 8.68GWKK314 pKa = 8.34SKK316 pKa = 9.54KK317 pKa = 8.6WAPARR322 pKa = 11.84AEE324 pKa = 4.01KK325 pKa = 10.63ALSEE329 pKa = 4.1LRR331 pKa = 11.84ATFAPKK337 pKa = 9.95FRR339 pKa = 11.84FDD341 pKa = 3.89AGIKK345 pKa = 10.11LEE347 pKa = 3.92PSKK350 pKa = 10.66RR351 pKa = 11.84GKK353 pKa = 10.44APRR356 pKa = 11.84LLIADD361 pKa = 4.13GDD363 pKa = 4.11RR364 pKa = 11.84GQVMSWVLIGTLEE377 pKa = 3.68AWLFKK382 pKa = 10.1RR383 pKa = 11.84WRR385 pKa = 11.84HH386 pKa = 5.12RR387 pKa = 11.84SIKK390 pKa = 10.29GLPKK394 pKa = 9.24TEE396 pKa = 3.5AMQRR400 pKa = 11.84VCQSLRR406 pKa = 11.84QKK408 pKa = 10.8DD409 pKa = 3.86PRR411 pKa = 11.84AGPDD415 pKa = 3.25TPDD418 pKa = 3.03IPVSIVEE425 pKa = 4.31NDD427 pKa = 3.07GSAWDD432 pKa = 3.56ACMSEE437 pKa = 4.35TLRR440 pKa = 11.84SLTEE444 pKa = 3.95NPVMEE449 pKa = 4.94AVAEE453 pKa = 4.1MVEE456 pKa = 4.4QYY458 pKa = 10.91FLVEE462 pKa = 4.61GPPEE466 pKa = 4.53FIDD469 pKa = 4.6ARR471 pKa = 11.84VASNRR476 pKa = 11.84LTKK479 pKa = 10.6LQLGFRR485 pKa = 11.84KK486 pKa = 10.22GKK488 pKa = 10.83GGDD491 pKa = 3.78DD492 pKa = 3.54YY493 pKa = 11.87VGKK496 pKa = 10.62CDD498 pKa = 4.75LPKK501 pKa = 10.85GKK503 pKa = 9.77AWQTIISAIRR513 pKa = 11.84RR514 pKa = 11.84SGCRR518 pKa = 11.84GTSCLNFLANMICWAWVIGGRR539 pKa = 11.84DD540 pKa = 3.15ACKK543 pKa = 10.03LVRR546 pKa = 11.84PQGARR551 pKa = 11.84VTCVDD556 pKa = 3.27GVVRR560 pKa = 11.84FVKK563 pKa = 10.01MVFEE567 pKa = 5.14GDD569 pKa = 3.5DD570 pKa = 4.02SILSFVGLNTAGQAADD586 pKa = 3.46QQLSRR591 pKa = 11.84EE592 pKa = 4.27YY593 pKa = 10.5IHH595 pKa = 6.99TCTQRR600 pKa = 11.84WTKK603 pKa = 10.44LGHH606 pKa = 6.02RR607 pKa = 11.84PKK609 pKa = 10.59LYY611 pKa = 9.05WRR613 pKa = 11.84AIGAVAEE620 pKa = 4.15FTGWHH625 pKa = 6.16FSVSDD630 pKa = 3.36VGIGSDD636 pKa = 4.01CAAPDD641 pKa = 4.74LIRR644 pKa = 11.84NLTNMAYY651 pKa = 10.14SINAAAINATAAGDD665 pKa = 3.83RR666 pKa = 11.84KK667 pKa = 10.9ALMSAVAPGVIARR680 pKa = 11.84LYY682 pKa = 10.21PMAEE686 pKa = 4.33KK687 pKa = 10.61FPMLCKK693 pKa = 10.51LLYY696 pKa = 10.58GQFAHH701 pKa = 7.18HH702 pKa = 6.78LRR704 pKa = 11.84DD705 pKa = 3.47TTTTDD710 pKa = 3.09LTRR713 pKa = 11.84DD714 pKa = 3.46EE715 pKa = 4.96IYY717 pKa = 10.93ALEE720 pKa = 4.77LEE722 pKa = 4.74PEE724 pKa = 4.07DD725 pKa = 5.15FGFKK729 pKa = 10.3EE730 pKa = 3.89SDD732 pKa = 3.54YY733 pKa = 10.7NTDD736 pKa = 2.69MDD738 pKa = 5.19LNIEE742 pKa = 4.12RR743 pKa = 11.84ATQRR747 pKa = 11.84FQPILEE753 pKa = 4.21RR754 pKa = 11.84FEE756 pKa = 3.98MAIGRR761 pKa = 11.84GNEE764 pKa = 3.79QEE766 pKa = 4.2EE767 pKa = 3.95ADD769 pKa = 3.82LAVRR773 pKa = 11.84LGLVPDD779 pKa = 3.39NDD781 pKa = 3.71KK782 pKa = 11.62YY783 pKa = 11.8YY784 pKa = 11.08DD785 pKa = 3.94LLDD788 pKa = 5.04AIEE791 pKa = 4.29GGYY794 pKa = 10.0RR795 pKa = 11.84VGADD799 pKa = 2.91SAAFARR805 pKa = 11.84SVEE808 pKa = 4.38AIRR811 pKa = 11.84EE812 pKa = 4.32GG813 pKa = 3.49
MM1 pKa = 7.4AVVSFRR7 pKa = 11.84PPSRR11 pKa = 11.84RR12 pKa = 11.84NDD14 pKa = 3.17LCYY17 pKa = 10.43EE18 pKa = 4.09CHH20 pKa = 6.38SWMLQHH26 pKa = 6.55LTYY29 pKa = 10.58EE30 pKa = 4.45HH31 pKa = 6.92ASGRR35 pKa = 11.84ADD37 pKa = 3.2VSDD40 pKa = 3.43TFLYY44 pKa = 10.54EE45 pKa = 5.1LEE47 pKa = 4.33LQVARR52 pKa = 11.84CGRR55 pKa = 11.84HH56 pKa = 5.52VEE58 pKa = 4.31TCAQRR63 pKa = 11.84IRR65 pKa = 11.84DD66 pKa = 3.41RR67 pKa = 11.84LGRR70 pKa = 11.84IKK72 pKa = 10.5HH73 pKa = 5.49ALCGQFHH80 pKa = 6.48GVGVHH85 pKa = 5.93HH86 pKa = 7.26VYY88 pKa = 9.12LTRR91 pKa = 11.84GEE93 pKa = 3.96VDD95 pKa = 2.47IMRR98 pKa = 11.84RR99 pKa = 11.84SGGAALYY106 pKa = 10.55SGAPTTTADD115 pKa = 3.86SNLHH119 pKa = 5.44FATYY123 pKa = 10.7RR124 pKa = 11.84LDD126 pKa = 3.46KK127 pKa = 10.66KK128 pKa = 9.56EE129 pKa = 3.87RR130 pKa = 11.84RR131 pKa = 11.84GDD133 pKa = 4.12PISCATIDD141 pKa = 3.6QLFGYY146 pKa = 9.73LAATIQQGRR155 pKa = 11.84VANRR159 pKa = 11.84HH160 pKa = 5.46CCCLHH165 pKa = 5.34TAGLGLRR172 pKa = 11.84NLRR175 pKa = 11.84PVKK178 pKa = 10.26RR179 pKa = 11.84MPEE182 pKa = 4.11DD183 pKa = 3.41EE184 pKa = 4.7PEE186 pKa = 4.26PPKK189 pKa = 10.96SDD191 pKa = 3.43DD192 pKa = 3.73GKK194 pKa = 10.42DD195 pKa = 3.57GKK197 pKa = 11.18VEE199 pKa = 4.29GPPEE203 pKa = 4.82DD204 pKa = 5.43DD205 pKa = 4.36PPQVDD210 pKa = 3.79CHH212 pKa = 6.11TCHH215 pKa = 6.82SGLAGIQLIHH225 pKa = 7.18DD226 pKa = 4.1VNEE229 pKa = 4.5GPEE232 pKa = 4.32TIPLAVRR239 pKa = 11.84YY240 pKa = 9.05LPQIEE245 pKa = 4.96EE246 pKa = 3.94RR247 pKa = 11.84LFWLNSQKK255 pKa = 10.66NVEE258 pKa = 4.1RR259 pKa = 11.84AIKK262 pKa = 9.85EE263 pKa = 4.18RR264 pKa = 11.84IEE266 pKa = 4.18EE267 pKa = 3.92PHH269 pKa = 6.83VPVTLSQEE277 pKa = 4.16EE278 pKa = 4.23KK279 pKa = 10.65DD280 pKa = 3.82DD281 pKa = 3.98LRR283 pKa = 11.84AVTQVMIDD291 pKa = 3.3QLKK294 pKa = 9.97ADD296 pKa = 4.47KK297 pKa = 11.16GLIDD301 pKa = 6.26KK302 pKa = 10.53IIQDD306 pKa = 3.55KK307 pKa = 10.98LGIYY311 pKa = 8.68GWKK314 pKa = 8.34SKK316 pKa = 9.54KK317 pKa = 8.6WAPARR322 pKa = 11.84AEE324 pKa = 4.01KK325 pKa = 10.63ALSEE329 pKa = 4.1LRR331 pKa = 11.84ATFAPKK337 pKa = 9.95FRR339 pKa = 11.84FDD341 pKa = 3.89AGIKK345 pKa = 10.11LEE347 pKa = 3.92PSKK350 pKa = 10.66RR351 pKa = 11.84GKK353 pKa = 10.44APRR356 pKa = 11.84LLIADD361 pKa = 4.13GDD363 pKa = 4.11RR364 pKa = 11.84GQVMSWVLIGTLEE377 pKa = 3.68AWLFKK382 pKa = 10.1RR383 pKa = 11.84WRR385 pKa = 11.84HH386 pKa = 5.12RR387 pKa = 11.84SIKK390 pKa = 10.29GLPKK394 pKa = 9.24TEE396 pKa = 3.5AMQRR400 pKa = 11.84VCQSLRR406 pKa = 11.84QKK408 pKa = 10.8DD409 pKa = 3.86PRR411 pKa = 11.84AGPDD415 pKa = 3.25TPDD418 pKa = 3.03IPVSIVEE425 pKa = 4.31NDD427 pKa = 3.07GSAWDD432 pKa = 3.56ACMSEE437 pKa = 4.35TLRR440 pKa = 11.84SLTEE444 pKa = 3.95NPVMEE449 pKa = 4.94AVAEE453 pKa = 4.1MVEE456 pKa = 4.4QYY458 pKa = 10.91FLVEE462 pKa = 4.61GPPEE466 pKa = 4.53FIDD469 pKa = 4.6ARR471 pKa = 11.84VASNRR476 pKa = 11.84LTKK479 pKa = 10.6LQLGFRR485 pKa = 11.84KK486 pKa = 10.22GKK488 pKa = 10.83GGDD491 pKa = 3.78DD492 pKa = 3.54YY493 pKa = 11.87VGKK496 pKa = 10.62CDD498 pKa = 4.75LPKK501 pKa = 10.85GKK503 pKa = 9.77AWQTIISAIRR513 pKa = 11.84RR514 pKa = 11.84SGCRR518 pKa = 11.84GTSCLNFLANMICWAWVIGGRR539 pKa = 11.84DD540 pKa = 3.15ACKK543 pKa = 10.03LVRR546 pKa = 11.84PQGARR551 pKa = 11.84VTCVDD556 pKa = 3.27GVVRR560 pKa = 11.84FVKK563 pKa = 10.01MVFEE567 pKa = 5.14GDD569 pKa = 3.5DD570 pKa = 4.02SILSFVGLNTAGQAADD586 pKa = 3.46QQLSRR591 pKa = 11.84EE592 pKa = 4.27YY593 pKa = 10.5IHH595 pKa = 6.99TCTQRR600 pKa = 11.84WTKK603 pKa = 10.44LGHH606 pKa = 6.02RR607 pKa = 11.84PKK609 pKa = 10.59LYY611 pKa = 9.05WRR613 pKa = 11.84AIGAVAEE620 pKa = 4.15FTGWHH625 pKa = 6.16FSVSDD630 pKa = 3.36VGIGSDD636 pKa = 4.01CAAPDD641 pKa = 4.74LIRR644 pKa = 11.84NLTNMAYY651 pKa = 10.14SINAAAINATAAGDD665 pKa = 3.83RR666 pKa = 11.84KK667 pKa = 10.9ALMSAVAPGVIARR680 pKa = 11.84LYY682 pKa = 10.21PMAEE686 pKa = 4.33KK687 pKa = 10.61FPMLCKK693 pKa = 10.51LLYY696 pKa = 10.58GQFAHH701 pKa = 7.18HH702 pKa = 6.78LRR704 pKa = 11.84DD705 pKa = 3.47TTTTDD710 pKa = 3.09LTRR713 pKa = 11.84DD714 pKa = 3.46EE715 pKa = 4.96IYY717 pKa = 10.93ALEE720 pKa = 4.77LEE722 pKa = 4.74PEE724 pKa = 4.07DD725 pKa = 5.15FGFKK729 pKa = 10.3EE730 pKa = 3.89SDD732 pKa = 3.54YY733 pKa = 10.7NTDD736 pKa = 2.69MDD738 pKa = 5.19LNIEE742 pKa = 4.12RR743 pKa = 11.84ATQRR747 pKa = 11.84FQPILEE753 pKa = 4.21RR754 pKa = 11.84FEE756 pKa = 3.98MAIGRR761 pKa = 11.84GNEE764 pKa = 3.79QEE766 pKa = 4.2EE767 pKa = 3.95ADD769 pKa = 3.82LAVRR773 pKa = 11.84LGLVPDD779 pKa = 3.39NDD781 pKa = 3.71KK782 pKa = 11.62YY783 pKa = 11.8YY784 pKa = 11.08DD785 pKa = 3.94LLDD788 pKa = 5.04AIEE791 pKa = 4.29GGYY794 pKa = 10.0RR795 pKa = 11.84VGADD799 pKa = 2.91SAAFARR805 pKa = 11.84SVEE808 pKa = 4.38AIRR811 pKa = 11.84EE812 pKa = 4.32GG813 pKa = 3.49
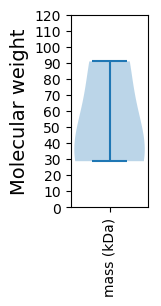
Molecular weight: 90.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL67|A0A1L3KL67_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 13 OX=1922742 PE=4 SV=1
MM1 pKa = 7.15VKK3 pKa = 10.34KK4 pKa = 10.23FQLKK8 pKa = 9.67KK9 pKa = 10.53KK10 pKa = 10.41RR11 pKa = 11.84VGQKK15 pKa = 9.92AATRR19 pKa = 11.84SKK21 pKa = 9.67ATRR24 pKa = 11.84VLAQGAGAVPRR35 pKa = 11.84KK36 pKa = 10.22AFGAGKK42 pKa = 9.96SLARR46 pKa = 11.84KK47 pKa = 8.09GWGMSMRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84MMEE59 pKa = 3.83GLNARR64 pKa = 11.84LPMHH68 pKa = 7.03LGLPRR73 pKa = 11.84PVGPYY78 pKa = 9.41QVIRR82 pKa = 11.84TTTLHH87 pKa = 4.95TTSAAVVIFAPLMRR101 pKa = 11.84DD102 pKa = 3.04ASTTSNGHH110 pKa = 5.24PVWYY114 pKa = 9.04HH115 pKa = 5.18ACGLEE120 pKa = 4.15DD121 pKa = 4.43AAPGPINGGAGTRR134 pKa = 11.84MISMPLDD141 pKa = 4.56GIGSAADD148 pKa = 3.38IVPAALTVQVMNKK161 pKa = 10.18ASLQQAEE168 pKa = 4.91GLFTMARR175 pKa = 11.84VSQQLALGGEE185 pKa = 4.15TRR187 pKa = 11.84TWEE190 pKa = 4.33DD191 pKa = 3.42FTHH194 pKa = 6.58EE195 pKa = 4.25FNSYY199 pKa = 9.86FKK201 pKa = 10.69PRR203 pKa = 11.84LLTGAKK209 pKa = 9.46LALRR213 pKa = 11.84GVTCNAVPINMNEE226 pKa = 3.9YY227 pKa = 10.22SDD229 pKa = 4.15FRR231 pKa = 11.84PGIPAISPAFGWYY244 pKa = 8.92TEE246 pKa = 4.13VAPAALSPIVFTQTGTPVDD265 pKa = 3.19ISFLVTIEE273 pKa = 3.47WRR275 pKa = 11.84VRR277 pKa = 11.84FDD279 pKa = 3.6PFHH282 pKa = 6.87PAAASHH288 pKa = 6.21TFHH291 pKa = 7.59PSTPDD296 pKa = 3.48GVWNSIIGAASAAGHH311 pKa = 5.69GVVDD315 pKa = 4.23IADD318 pKa = 3.75EE319 pKa = 4.18VADD322 pKa = 3.86VGFDD326 pKa = 3.29AAAVAGVAAALL337 pKa = 3.79
MM1 pKa = 7.15VKK3 pKa = 10.34KK4 pKa = 10.23FQLKK8 pKa = 9.67KK9 pKa = 10.53KK10 pKa = 10.41RR11 pKa = 11.84VGQKK15 pKa = 9.92AATRR19 pKa = 11.84SKK21 pKa = 9.67ATRR24 pKa = 11.84VLAQGAGAVPRR35 pKa = 11.84KK36 pKa = 10.22AFGAGKK42 pKa = 9.96SLARR46 pKa = 11.84KK47 pKa = 8.09GWGMSMRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84MMEE59 pKa = 3.83GLNARR64 pKa = 11.84LPMHH68 pKa = 7.03LGLPRR73 pKa = 11.84PVGPYY78 pKa = 9.41QVIRR82 pKa = 11.84TTTLHH87 pKa = 4.95TTSAAVVIFAPLMRR101 pKa = 11.84DD102 pKa = 3.04ASTTSNGHH110 pKa = 5.24PVWYY114 pKa = 9.04HH115 pKa = 5.18ACGLEE120 pKa = 4.15DD121 pKa = 4.43AAPGPINGGAGTRR134 pKa = 11.84MISMPLDD141 pKa = 4.56GIGSAADD148 pKa = 3.38IVPAALTVQVMNKK161 pKa = 10.18ASLQQAEE168 pKa = 4.91GLFTMARR175 pKa = 11.84VSQQLALGGEE185 pKa = 4.15TRR187 pKa = 11.84TWEE190 pKa = 4.33DD191 pKa = 3.42FTHH194 pKa = 6.58EE195 pKa = 4.25FNSYY199 pKa = 9.86FKK201 pKa = 10.69PRR203 pKa = 11.84LLTGAKK209 pKa = 9.46LALRR213 pKa = 11.84GVTCNAVPINMNEE226 pKa = 3.9YY227 pKa = 10.22SDD229 pKa = 4.15FRR231 pKa = 11.84PGIPAISPAFGWYY244 pKa = 8.92TEE246 pKa = 4.13VAPAALSPIVFTQTGTPVDD265 pKa = 3.19ISFLVTIEE273 pKa = 3.47WRR275 pKa = 11.84VRR277 pKa = 11.84FDD279 pKa = 3.6PFHH282 pKa = 6.87PAAASHH288 pKa = 6.21TFHH291 pKa = 7.59PSTPDD296 pKa = 3.48GVWNSIIGAASAAGHH311 pKa = 5.69GVVDD315 pKa = 4.23IADD318 pKa = 3.75EE319 pKa = 4.18VADD322 pKa = 3.86VGFDD326 pKa = 3.29AAAVAGVAAALL337 pKa = 3.79
Molecular weight: 35.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1406 |
256 |
813 |
468.7 |
51.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.597 ± 1.474 | 2.418 ± 0.679 |
6.046 ± 0.841 | 5.477 ± 0.937 |
3.912 ± 0.622 | 8.748 ± 0.749 |
3.129 ± 0.735 | 4.339 ± 1.042 |
4.054 ± 0.991 | 7.397 ± 1.388 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.373 | 2.418 ± 0.266 |
5.05 ± 0.734 | 3.841 ± 0.422 |
7.895 ± 0.768 | 5.974 ± 1.484 |
5.334 ± 0.621 | 6.543 ± 0.52 |
1.991 ± 0.357 | 2.347 ± 0.3 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
