
Moheibacter sediminis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Weeksellaceae; Moheibacter
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
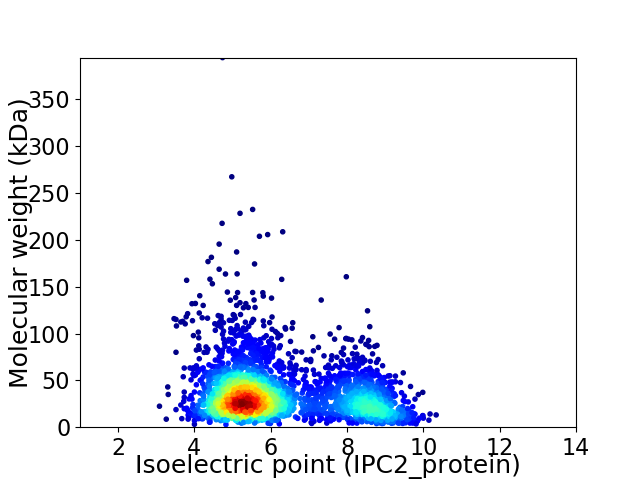
Virtual 2D-PAGE plot for 2974 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W2C911|A0A1W2C911_9FLAO Putative fluoride ion transporter CrcB OS=Moheibacter sediminis OX=1434700 GN=crcB PE=3 SV=1
MM1 pKa = 7.82KK2 pKa = 9.9IFNYY6 pKa = 9.37ILSFIFISGLITSCSDD22 pKa = 4.21DD23 pKa = 5.53DD24 pKa = 4.0FTKK27 pKa = 10.89TNLNDD32 pKa = 3.63ALKK35 pKa = 10.42PSITSPSEE43 pKa = 3.66GSSYY47 pKa = 11.43EE48 pKa = 3.92LLQEE52 pKa = 4.2NAGNEE57 pKa = 4.1AFTMEE62 pKa = 4.15WEE64 pKa = 4.15EE65 pKa = 4.26ADD67 pKa = 3.9FGVNSPVKK75 pKa = 10.3YY76 pKa = 10.16DD77 pKa = 3.55VQASSSEE84 pKa = 4.18DD85 pKa = 3.48FSEE88 pKa = 4.22TTNVITALTGTSTSLTVGRR107 pKa = 11.84LNAVSLSSGLPFDD120 pKa = 4.23VEE122 pKa = 4.31GNLFIRR128 pKa = 11.84VKK130 pKa = 10.61AYY132 pKa = 10.77LGVAGSSGFIYY143 pKa = 10.37SEE145 pKa = 4.58PISLNVTPYY154 pKa = 10.84EE155 pKa = 3.92PLIDD159 pKa = 5.57LSTPWGMVGSAVPNGWEE176 pKa = 4.41GPDD179 pKa = 3.31VPFWKK184 pKa = 9.47TDD186 pKa = 3.53LNGIDD191 pKa = 3.56DD192 pKa = 4.72GKK194 pKa = 10.3YY195 pKa = 9.18VAYY198 pKa = 8.6ATLTDD203 pKa = 3.62GDD205 pKa = 3.64IKK207 pKa = 10.61IRR209 pKa = 11.84KK210 pKa = 8.55DD211 pKa = 3.45NSWDD215 pKa = 3.49EE216 pKa = 4.38NYY218 pKa = 10.87GGAAGTLVSNGDD230 pKa = 4.95DD231 pKa = 3.27IPMTAGTYY239 pKa = 10.1KK240 pKa = 10.73FNVDD244 pKa = 3.6MNALTYY250 pKa = 10.47EE251 pKa = 4.14FEE253 pKa = 4.3EE254 pKa = 4.59YY255 pKa = 10.35SWGIVGDD262 pKa = 3.8ATPNGWNGPDD272 pKa = 3.42VQLQYY277 pKa = 11.52NGATNTWDD285 pKa = 3.61AEE287 pKa = 4.54VEE289 pKa = 4.28FTNGNIKK296 pKa = 10.19FRR298 pKa = 11.84FNNAWALSYY307 pKa = 11.4GDD309 pKa = 3.82TGADD313 pKa = 3.07GTLEE317 pKa = 4.19SEE319 pKa = 4.38NGADD323 pKa = 3.75IPVTAGNYY331 pKa = 8.8LVSVDD336 pKa = 4.41FTNLTYY342 pKa = 10.68SITPLL347 pKa = 3.45
MM1 pKa = 7.82KK2 pKa = 9.9IFNYY6 pKa = 9.37ILSFIFISGLITSCSDD22 pKa = 4.21DD23 pKa = 5.53DD24 pKa = 4.0FTKK27 pKa = 10.89TNLNDD32 pKa = 3.63ALKK35 pKa = 10.42PSITSPSEE43 pKa = 3.66GSSYY47 pKa = 11.43EE48 pKa = 3.92LLQEE52 pKa = 4.2NAGNEE57 pKa = 4.1AFTMEE62 pKa = 4.15WEE64 pKa = 4.15EE65 pKa = 4.26ADD67 pKa = 3.9FGVNSPVKK75 pKa = 10.3YY76 pKa = 10.16DD77 pKa = 3.55VQASSSEE84 pKa = 4.18DD85 pKa = 3.48FSEE88 pKa = 4.22TTNVITALTGTSTSLTVGRR107 pKa = 11.84LNAVSLSSGLPFDD120 pKa = 4.23VEE122 pKa = 4.31GNLFIRR128 pKa = 11.84VKK130 pKa = 10.61AYY132 pKa = 10.77LGVAGSSGFIYY143 pKa = 10.37SEE145 pKa = 4.58PISLNVTPYY154 pKa = 10.84EE155 pKa = 3.92PLIDD159 pKa = 5.57LSTPWGMVGSAVPNGWEE176 pKa = 4.41GPDD179 pKa = 3.31VPFWKK184 pKa = 9.47TDD186 pKa = 3.53LNGIDD191 pKa = 3.56DD192 pKa = 4.72GKK194 pKa = 10.3YY195 pKa = 9.18VAYY198 pKa = 8.6ATLTDD203 pKa = 3.62GDD205 pKa = 3.64IKK207 pKa = 10.61IRR209 pKa = 11.84KK210 pKa = 8.55DD211 pKa = 3.45NSWDD215 pKa = 3.49EE216 pKa = 4.38NYY218 pKa = 10.87GGAAGTLVSNGDD230 pKa = 4.95DD231 pKa = 3.27IPMTAGTYY239 pKa = 10.1KK240 pKa = 10.73FNVDD244 pKa = 3.6MNALTYY250 pKa = 10.47EE251 pKa = 4.14FEE253 pKa = 4.3EE254 pKa = 4.59YY255 pKa = 10.35SWGIVGDD262 pKa = 3.8ATPNGWNGPDD272 pKa = 3.42VQLQYY277 pKa = 11.52NGATNTWDD285 pKa = 3.61AEE287 pKa = 4.54VEE289 pKa = 4.28FTNGNIKK296 pKa = 10.19FRR298 pKa = 11.84FNNAWALSYY307 pKa = 11.4GDD309 pKa = 3.82TGADD313 pKa = 3.07GTLEE317 pKa = 4.19SEE319 pKa = 4.38NGADD323 pKa = 3.75IPVTAGNYY331 pKa = 8.8LVSVDD336 pKa = 4.41FTNLTYY342 pKa = 10.68SITPLL347 pKa = 3.45
Molecular weight: 37.52 kDa
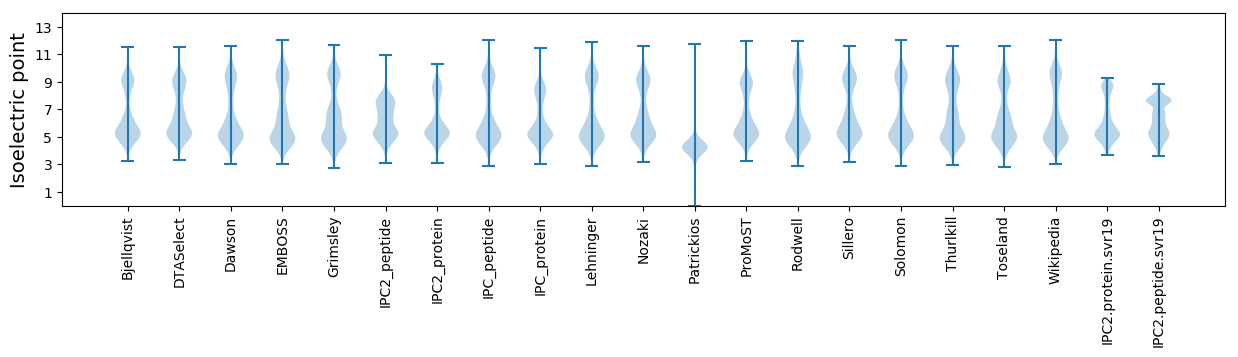
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W2ATL0|A0A1W2ATL0_9FLAO Putative iron-regulated protein OS=Moheibacter sediminis OX=1434700 GN=SAMN06296427_10541 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84IVNNFDD22 pKa = 3.19NVEE25 pKa = 4.17SVKK28 pKa = 10.43PEE30 pKa = 3.49KK31 pKa = 10.78SLTVGKK37 pKa = 10.15KK38 pKa = 9.23RR39 pKa = 11.84SGGRR43 pKa = 11.84NHH45 pKa = 6.53SGKK48 pKa = 8.3MTMRR52 pKa = 11.84YY53 pKa = 9.23IGGGHH58 pKa = 5.49KK59 pKa = 9.73QSYY62 pKa = 9.64RR63 pKa = 11.84IVDD66 pKa = 4.1FKK68 pKa = 11.27RR69 pKa = 11.84EE70 pKa = 4.05KK71 pKa = 10.74EE72 pKa = 4.26GVATVDD78 pKa = 4.3SIQYY82 pKa = 10.04DD83 pKa = 3.71PNRR86 pKa = 11.84TAFIALIVYY95 pKa = 10.33GDD97 pKa = 3.55GEE99 pKa = 4.33KK100 pKa = 10.32RR101 pKa = 11.84YY102 pKa = 10.71VIAQNGLKK110 pKa = 10.12KK111 pKa = 10.73GQTIEE116 pKa = 4.0SGEE119 pKa = 4.08NVAPEE124 pKa = 3.74IGNTLKK130 pKa = 10.95LKK132 pKa = 10.04NVPLGTVISCIEE144 pKa = 3.9LRR146 pKa = 11.84PGQGALMARR155 pKa = 11.84SAGSYY160 pKa = 9.0AQLVARR166 pKa = 11.84DD167 pKa = 3.78GKK169 pKa = 10.37YY170 pKa = 9.12ATVKK174 pKa = 10.47LPSGEE179 pKa = 3.88SRR181 pKa = 11.84MILVEE186 pKa = 4.29CKK188 pKa = 9.45ATIGVVSNSDD198 pKa = 3.13HH199 pKa = 5.64QLEE202 pKa = 4.55VSGKK206 pKa = 10.07AGRR209 pKa = 11.84TRR211 pKa = 11.84WKK213 pKa = 9.99GRR215 pKa = 11.84RR216 pKa = 11.84PRR218 pKa = 11.84TRR220 pKa = 11.84PMVMNPVDD228 pKa = 3.52HH229 pKa = 7.07PMGGGEE235 pKa = 3.92GRR237 pKa = 11.84ATGGIPRR244 pKa = 11.84SRR246 pKa = 11.84NGQPSKK252 pKa = 10.7GYY254 pKa = 8.28KK255 pKa = 8.33TRR257 pKa = 11.84APKK260 pKa = 8.88KK261 pKa = 7.78TSSKK265 pKa = 10.57YY266 pKa = 8.93IVEE269 pKa = 4.17RR270 pKa = 11.84RR271 pKa = 11.84KK272 pKa = 10.24KK273 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84IVNNFDD22 pKa = 3.19NVEE25 pKa = 4.17SVKK28 pKa = 10.43PEE30 pKa = 3.49KK31 pKa = 10.78SLTVGKK37 pKa = 10.15KK38 pKa = 9.23RR39 pKa = 11.84SGGRR43 pKa = 11.84NHH45 pKa = 6.53SGKK48 pKa = 8.3MTMRR52 pKa = 11.84YY53 pKa = 9.23IGGGHH58 pKa = 5.49KK59 pKa = 9.73QSYY62 pKa = 9.64RR63 pKa = 11.84IVDD66 pKa = 4.1FKK68 pKa = 11.27RR69 pKa = 11.84EE70 pKa = 4.05KK71 pKa = 10.74EE72 pKa = 4.26GVATVDD78 pKa = 4.3SIQYY82 pKa = 10.04DD83 pKa = 3.71PNRR86 pKa = 11.84TAFIALIVYY95 pKa = 10.33GDD97 pKa = 3.55GEE99 pKa = 4.33KK100 pKa = 10.32RR101 pKa = 11.84YY102 pKa = 10.71VIAQNGLKK110 pKa = 10.12KK111 pKa = 10.73GQTIEE116 pKa = 4.0SGEE119 pKa = 4.08NVAPEE124 pKa = 3.74IGNTLKK130 pKa = 10.95LKK132 pKa = 10.04NVPLGTVISCIEE144 pKa = 3.9LRR146 pKa = 11.84PGQGALMARR155 pKa = 11.84SAGSYY160 pKa = 9.0AQLVARR166 pKa = 11.84DD167 pKa = 3.78GKK169 pKa = 10.37YY170 pKa = 9.12ATVKK174 pKa = 10.47LPSGEE179 pKa = 3.88SRR181 pKa = 11.84MILVEE186 pKa = 4.29CKK188 pKa = 9.45ATIGVVSNSDD198 pKa = 3.13HH199 pKa = 5.64QLEE202 pKa = 4.55VSGKK206 pKa = 10.07AGRR209 pKa = 11.84TRR211 pKa = 11.84WKK213 pKa = 9.99GRR215 pKa = 11.84RR216 pKa = 11.84PRR218 pKa = 11.84TRR220 pKa = 11.84PMVMNPVDD228 pKa = 3.52HH229 pKa = 7.07PMGGGEE235 pKa = 3.92GRR237 pKa = 11.84ATGGIPRR244 pKa = 11.84SRR246 pKa = 11.84NGQPSKK252 pKa = 10.7GYY254 pKa = 8.28KK255 pKa = 8.33TRR257 pKa = 11.84APKK260 pKa = 8.88KK261 pKa = 7.78TSSKK265 pKa = 10.57YY266 pKa = 8.93IVEE269 pKa = 4.17RR270 pKa = 11.84RR271 pKa = 11.84KK272 pKa = 10.24KK273 pKa = 10.1
Molecular weight: 29.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
985835 |
28 |
3536 |
331.5 |
37.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.889 ± 0.047 | 0.7 ± 0.015 |
5.413 ± 0.035 | 7.13 ± 0.046 |
5.412 ± 0.033 | 6.498 ± 0.049 |
1.696 ± 0.019 | 8.204 ± 0.043 |
7.651 ± 0.066 | 9.064 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.28 ± 0.02 | 6.588 ± 0.048 |
3.323 ± 0.027 | 3.623 ± 0.022 |
3.223 ± 0.029 | 6.646 ± 0.037 |
5.513 ± 0.045 | 5.926 ± 0.036 |
1.069 ± 0.017 | 4.152 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
