
Cucumis melo var. makuwa
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis; Cucumis melo; Cucumi
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
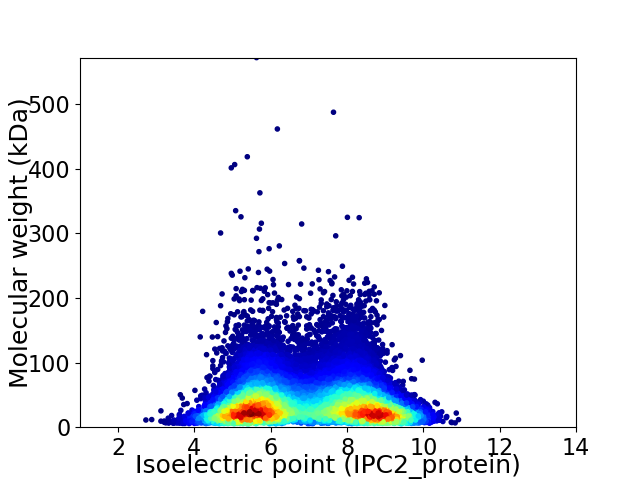
Virtual 2D-PAGE plot for 37791 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5A7THJ8|A0A5A7THJ8_CUCME Pol protein OS=Cucumis melo var. makuwa OX=1194695 GN=E5676_scaffold205G001980 PE=4 SV=1
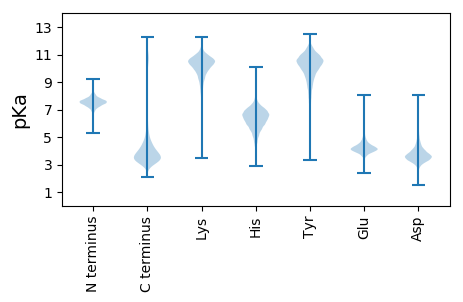
MM1 pKa = 7.56FLTIHH6 pKa = 7.29LILPLKK12 pKa = 9.57WEE14 pKa = 4.56AYY16 pKa = 3.97WTKK19 pKa = 10.96LMSCPQYY26 pKa = 10.62VDD28 pKa = 4.09LRR30 pKa = 11.84IISCEE35 pKa = 3.78QEE37 pKa = 3.82FTVCSRR43 pKa = 11.84LRR45 pKa = 11.84LSYY48 pKa = 10.42LVKK51 pKa = 10.65DD52 pKa = 4.03DD53 pKa = 4.46LLDD56 pKa = 3.95YY57 pKa = 11.23GHH59 pKa = 7.25IEE61 pKa = 4.18YY62 pKa = 10.83EE63 pKa = 4.19EE64 pKa = 5.78GDD66 pKa = 4.09DD67 pKa = 4.14PFGSAAFKK75 pKa = 10.32MKK77 pKa = 10.32IKK79 pKa = 10.36IILSTATILKK89 pKa = 9.09TIEE92 pKa = 3.96EE93 pKa = 4.42EE94 pKa = 4.1NEE96 pKa = 3.75KK97 pKa = 11.12DD98 pKa = 5.26LFSCDD103 pKa = 3.76YY104 pKa = 11.05IEE106 pKa = 6.51DD107 pKa = 3.64EE108 pKa = 5.75DD109 pKa = 4.79NPLDD113 pKa = 4.15CGSIEE118 pKa = 5.98DD119 pKa = 4.39YY120 pKa = 11.52DD121 pKa = 4.33EE122 pKa = 6.51DD123 pKa = 6.1DD124 pKa = 4.42PFCCGRR130 pKa = 11.84IEE132 pKa = 5.82DD133 pKa = 4.04EE134 pKa = 5.08DD135 pKa = 5.81GYY137 pKa = 11.62DD138 pKa = 4.38PLCCDD143 pKa = 5.29DD144 pKa = 6.45LLDD147 pKa = 4.11CSRR150 pKa = 11.84NEE152 pKa = 5.42DD153 pKa = 3.8EE154 pKa = 5.82NKK156 pKa = 10.63DD157 pKa = 3.44DD158 pKa = 4.23ALNYY162 pKa = 10.95GCMEE166 pKa = 5.0DD167 pKa = 4.8KK168 pKa = 10.71DD169 pKa = 5.18DD170 pKa = 4.69PFDD173 pKa = 3.88YY174 pKa = 11.19SHH176 pKa = 8.15IEE178 pKa = 5.2DD179 pKa = 4.45EE180 pKa = 6.06DD181 pKa = 4.15EE182 pKa = 5.26DD183 pKa = 5.29DD184 pKa = 4.05PLGYY188 pKa = 10.7VCIEE192 pKa = 5.04DD193 pKa = 4.06EE194 pKa = 6.02DD195 pKa = 4.67EE196 pKa = 6.36DD197 pKa = 4.83DD198 pKa = 6.35LLDD201 pKa = 4.02EE202 pKa = 4.75DD203 pKa = 5.37HH204 pKa = 7.6PLGCDD209 pKa = 3.44CIEE212 pKa = 4.85NKK214 pKa = 10.54DD215 pKa = 3.98DD216 pKa = 6.05DD217 pKa = 5.44DD218 pKa = 4.99PLICDD223 pKa = 4.66RR224 pKa = 11.84IKK226 pKa = 11.35DD227 pKa = 3.93EE228 pKa = 6.76DD229 pKa = 3.87DD230 pKa = 4.41LFYY233 pKa = 10.95CGCIEE238 pKa = 6.01DD239 pKa = 4.25EE240 pKa = 5.36DD241 pKa = 4.61EE242 pKa = 5.81DD243 pKa = 5.14GPLDD247 pKa = 3.93FVLIKK252 pKa = 10.93DD253 pKa = 3.72KK254 pKa = 11.55DD255 pKa = 3.95EE256 pKa = 4.64NDD258 pKa = 3.72PLSCGRR264 pKa = 11.84IEE266 pKa = 4.53VEE268 pKa = 5.03FDD270 pKa = 3.25PFCYY274 pKa = 10.13DD275 pKa = 4.38HH276 pKa = 7.08EE277 pKa = 5.38GKK279 pKa = 10.4DD280 pKa = 3.41EE281 pKa = 5.24DD282 pKa = 5.42DD283 pKa = 4.06PFGCRR288 pKa = 11.84RR289 pKa = 11.84IDD291 pKa = 4.66DD292 pKa = 3.85EE293 pKa = 6.42HH294 pKa = 7.6EE295 pKa = 4.34DD296 pKa = 4.25ASLNLDD302 pKa = 4.14NIKK305 pKa = 10.89DD306 pKa = 3.76EE307 pKa = 4.43YY308 pKa = 11.54DD309 pKa = 3.43PLDD312 pKa = 3.49YY313 pKa = 11.24DD314 pKa = 4.41RR315 pKa = 11.84IEE317 pKa = 6.06DD318 pKa = 3.94EE319 pKa = 6.51DD320 pKa = 4.69EE321 pKa = 5.07DD322 pKa = 5.13DD323 pKa = 4.61PLNIDD328 pKa = 3.58RR329 pKa = 11.84SEE331 pKa = 4.67DD332 pKa = 3.52KK333 pKa = 10.94DD334 pKa = 4.66DD335 pKa = 4.12NDD337 pKa = 4.17PLGFRR342 pKa = 11.84CIKK345 pKa = 10.95DD346 pKa = 3.65EE347 pKa = 4.37VDD349 pKa = 3.23DD350 pKa = 3.86TLGFGRR356 pKa = 11.84IKK358 pKa = 11.1DD359 pKa = 4.02EE360 pKa = 4.9DD361 pKa = 4.15DD362 pKa = 5.06HH363 pKa = 8.79PLDD366 pKa = 4.08CNKK369 pKa = 9.94WEE371 pKa = 4.04MLKK374 pKa = 10.95LLACNISMRR383 pKa = 11.84SRR385 pKa = 11.84IQQ387 pKa = 2.94
MM1 pKa = 7.56FLTIHH6 pKa = 7.29LILPLKK12 pKa = 9.57WEE14 pKa = 4.56AYY16 pKa = 3.97WTKK19 pKa = 10.96LMSCPQYY26 pKa = 10.62VDD28 pKa = 4.09LRR30 pKa = 11.84IISCEE35 pKa = 3.78QEE37 pKa = 3.82FTVCSRR43 pKa = 11.84LRR45 pKa = 11.84LSYY48 pKa = 10.42LVKK51 pKa = 10.65DD52 pKa = 4.03DD53 pKa = 4.46LLDD56 pKa = 3.95YY57 pKa = 11.23GHH59 pKa = 7.25IEE61 pKa = 4.18YY62 pKa = 10.83EE63 pKa = 4.19EE64 pKa = 5.78GDD66 pKa = 4.09DD67 pKa = 4.14PFGSAAFKK75 pKa = 10.32MKK77 pKa = 10.32IKK79 pKa = 10.36IILSTATILKK89 pKa = 9.09TIEE92 pKa = 3.96EE93 pKa = 4.42EE94 pKa = 4.1NEE96 pKa = 3.75KK97 pKa = 11.12DD98 pKa = 5.26LFSCDD103 pKa = 3.76YY104 pKa = 11.05IEE106 pKa = 6.51DD107 pKa = 3.64EE108 pKa = 5.75DD109 pKa = 4.79NPLDD113 pKa = 4.15CGSIEE118 pKa = 5.98DD119 pKa = 4.39YY120 pKa = 11.52DD121 pKa = 4.33EE122 pKa = 6.51DD123 pKa = 6.1DD124 pKa = 4.42PFCCGRR130 pKa = 11.84IEE132 pKa = 5.82DD133 pKa = 4.04EE134 pKa = 5.08DD135 pKa = 5.81GYY137 pKa = 11.62DD138 pKa = 4.38PLCCDD143 pKa = 5.29DD144 pKa = 6.45LLDD147 pKa = 4.11CSRR150 pKa = 11.84NEE152 pKa = 5.42DD153 pKa = 3.8EE154 pKa = 5.82NKK156 pKa = 10.63DD157 pKa = 3.44DD158 pKa = 4.23ALNYY162 pKa = 10.95GCMEE166 pKa = 5.0DD167 pKa = 4.8KK168 pKa = 10.71DD169 pKa = 5.18DD170 pKa = 4.69PFDD173 pKa = 3.88YY174 pKa = 11.19SHH176 pKa = 8.15IEE178 pKa = 5.2DD179 pKa = 4.45EE180 pKa = 6.06DD181 pKa = 4.15EE182 pKa = 5.26DD183 pKa = 5.29DD184 pKa = 4.05PLGYY188 pKa = 10.7VCIEE192 pKa = 5.04DD193 pKa = 4.06EE194 pKa = 6.02DD195 pKa = 4.67EE196 pKa = 6.36DD197 pKa = 4.83DD198 pKa = 6.35LLDD201 pKa = 4.02EE202 pKa = 4.75DD203 pKa = 5.37HH204 pKa = 7.6PLGCDD209 pKa = 3.44CIEE212 pKa = 4.85NKK214 pKa = 10.54DD215 pKa = 3.98DD216 pKa = 6.05DD217 pKa = 5.44DD218 pKa = 4.99PLICDD223 pKa = 4.66RR224 pKa = 11.84IKK226 pKa = 11.35DD227 pKa = 3.93EE228 pKa = 6.76DD229 pKa = 3.87DD230 pKa = 4.41LFYY233 pKa = 10.95CGCIEE238 pKa = 6.01DD239 pKa = 4.25EE240 pKa = 5.36DD241 pKa = 4.61EE242 pKa = 5.81DD243 pKa = 5.14GPLDD247 pKa = 3.93FVLIKK252 pKa = 10.93DD253 pKa = 3.72KK254 pKa = 11.55DD255 pKa = 3.95EE256 pKa = 4.64NDD258 pKa = 3.72PLSCGRR264 pKa = 11.84IEE266 pKa = 4.53VEE268 pKa = 5.03FDD270 pKa = 3.25PFCYY274 pKa = 10.13DD275 pKa = 4.38HH276 pKa = 7.08EE277 pKa = 5.38GKK279 pKa = 10.4DD280 pKa = 3.41EE281 pKa = 5.24DD282 pKa = 5.42DD283 pKa = 4.06PFGCRR288 pKa = 11.84RR289 pKa = 11.84IDD291 pKa = 4.66DD292 pKa = 3.85EE293 pKa = 6.42HH294 pKa = 7.6EE295 pKa = 4.34DD296 pKa = 4.25ASLNLDD302 pKa = 4.14NIKK305 pKa = 10.89DD306 pKa = 3.76EE307 pKa = 4.43YY308 pKa = 11.54DD309 pKa = 3.43PLDD312 pKa = 3.49YY313 pKa = 11.24DD314 pKa = 4.41RR315 pKa = 11.84IEE317 pKa = 6.06DD318 pKa = 3.94EE319 pKa = 6.51DD320 pKa = 4.69EE321 pKa = 5.07DD322 pKa = 5.13DD323 pKa = 4.61PLNIDD328 pKa = 3.58RR329 pKa = 11.84SEE331 pKa = 4.67DD332 pKa = 3.52KK333 pKa = 10.94DD334 pKa = 4.66DD335 pKa = 4.12NDD337 pKa = 4.17PLGFRR342 pKa = 11.84CIKK345 pKa = 10.95DD346 pKa = 3.65EE347 pKa = 4.37VDD349 pKa = 3.23DD350 pKa = 3.86TLGFGRR356 pKa = 11.84IKK358 pKa = 11.1DD359 pKa = 4.02EE360 pKa = 4.9DD361 pKa = 4.15DD362 pKa = 5.06HH363 pKa = 8.79PLDD366 pKa = 4.08CNKK369 pKa = 9.94WEE371 pKa = 4.04MLKK374 pKa = 10.95LLACNISMRR383 pKa = 11.84SRR385 pKa = 11.84IQQ387 pKa = 2.94
Molecular weight: 45.1 kDa
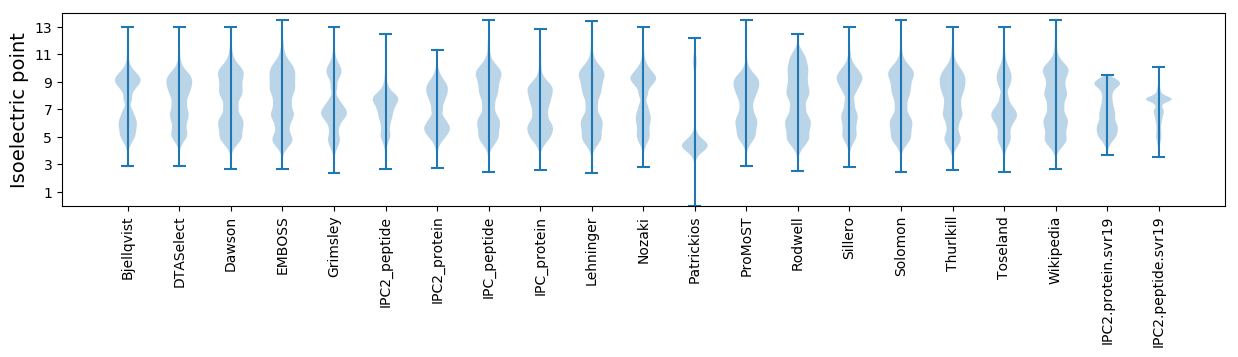
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5A7SN37|A0A5A7SN37_CUCME Uncharacterized protein OS=Cucumis melo var. makuwa OX=1194695 GN=E6C27_scaffold581G00420 PE=4 SV=1
MM1 pKa = 7.57RR2 pKa = 11.84ASFKK6 pKa = 9.38STRR9 pKa = 11.84LMRR12 pKa = 11.84ASFGSTRR19 pKa = 11.84LMRR22 pKa = 11.84ASFGSTRR29 pKa = 11.84LMRR32 pKa = 11.84ASFGSTRR39 pKa = 11.84LMRR42 pKa = 11.84ASFGSTRR49 pKa = 11.84LMRR52 pKa = 11.84ASFGSTRR59 pKa = 11.84LMRR62 pKa = 11.84ASFGSTRR69 pKa = 11.84LMRR72 pKa = 11.84ASFGSTRR79 pKa = 11.84LMRR82 pKa = 11.84ASFGSTRR89 pKa = 11.84LMRR92 pKa = 11.84ASFGSTRR99 pKa = 11.84LMRR102 pKa = 11.84ASFGSTRR109 pKa = 11.84LMRR112 pKa = 11.84ASFGSTRR119 pKa = 11.84LMRR122 pKa = 11.84ASFGSTRR129 pKa = 11.84LMRR132 pKa = 11.84ASFGSTRR139 pKa = 11.84LMRR142 pKa = 11.84ASFGSTRR149 pKa = 11.84LMRR152 pKa = 11.84ASFGSTRR159 pKa = 11.84LMRR162 pKa = 11.84ASFGSTRR169 pKa = 11.84LMRR172 pKa = 11.84ASFGSTRR179 pKa = 11.84LMRR182 pKa = 11.84ASFGSTRR189 pKa = 11.84LMRR192 pKa = 11.84ASFGSTRR199 pKa = 11.84LMRR202 pKa = 11.84ASFGSTRR209 pKa = 11.84LMRR212 pKa = 11.84ASFGSTRR219 pKa = 11.84LMRR222 pKa = 11.84ASFGSTRR229 pKa = 11.84LMRR232 pKa = 11.84ASFGSTRR239 pKa = 11.84LICASFGSTRR249 pKa = 11.84LICAFYY255 pKa = 8.81GTTRR259 pKa = 11.84LLFKK263 pKa = 9.78GTARR267 pKa = 11.84GRR269 pKa = 11.84PTRR272 pKa = 11.84GRR274 pKa = 11.84KK275 pKa = 8.91DD276 pKa = 2.97AA277 pKa = 4.69
MM1 pKa = 7.57RR2 pKa = 11.84ASFKK6 pKa = 9.38STRR9 pKa = 11.84LMRR12 pKa = 11.84ASFGSTRR19 pKa = 11.84LMRR22 pKa = 11.84ASFGSTRR29 pKa = 11.84LMRR32 pKa = 11.84ASFGSTRR39 pKa = 11.84LMRR42 pKa = 11.84ASFGSTRR49 pKa = 11.84LMRR52 pKa = 11.84ASFGSTRR59 pKa = 11.84LMRR62 pKa = 11.84ASFGSTRR69 pKa = 11.84LMRR72 pKa = 11.84ASFGSTRR79 pKa = 11.84LMRR82 pKa = 11.84ASFGSTRR89 pKa = 11.84LMRR92 pKa = 11.84ASFGSTRR99 pKa = 11.84LMRR102 pKa = 11.84ASFGSTRR109 pKa = 11.84LMRR112 pKa = 11.84ASFGSTRR119 pKa = 11.84LMRR122 pKa = 11.84ASFGSTRR129 pKa = 11.84LMRR132 pKa = 11.84ASFGSTRR139 pKa = 11.84LMRR142 pKa = 11.84ASFGSTRR149 pKa = 11.84LMRR152 pKa = 11.84ASFGSTRR159 pKa = 11.84LMRR162 pKa = 11.84ASFGSTRR169 pKa = 11.84LMRR172 pKa = 11.84ASFGSTRR179 pKa = 11.84LMRR182 pKa = 11.84ASFGSTRR189 pKa = 11.84LMRR192 pKa = 11.84ASFGSTRR199 pKa = 11.84LMRR202 pKa = 11.84ASFGSTRR209 pKa = 11.84LMRR212 pKa = 11.84ASFGSTRR219 pKa = 11.84LMRR222 pKa = 11.84ASFGSTRR229 pKa = 11.84LMRR232 pKa = 11.84ASFGSTRR239 pKa = 11.84LICASFGSTRR249 pKa = 11.84LICAFYY255 pKa = 8.81GTTRR259 pKa = 11.84LLFKK263 pKa = 9.78GTARR267 pKa = 11.84GRR269 pKa = 11.84PTRR272 pKa = 11.84GRR274 pKa = 11.84KK275 pKa = 8.91DD276 pKa = 2.97AA277 pKa = 4.69
Molecular weight: 30.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14011924 |
35 |
5438 |
370.8 |
41.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.215 ± 0.011 | 1.849 ± 0.006 |
5.333 ± 0.009 | 6.63 ± 0.015 |
4.336 ± 0.009 | 6.03 ± 0.01 |
2.451 ± 0.005 | 5.433 ± 0.01 |
6.504 ± 0.014 | 9.505 ± 0.012 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.006 | 4.327 ± 0.01 |
4.822 ± 0.011 | 3.712 ± 0.008 |
5.726 ± 0.01 | 8.616 ± 0.014 |
5.111 ± 0.008 | 6.688 ± 0.01 |
1.351 ± 0.004 | 2.857 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
