
Pistachio ampelovirus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Ampelovirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
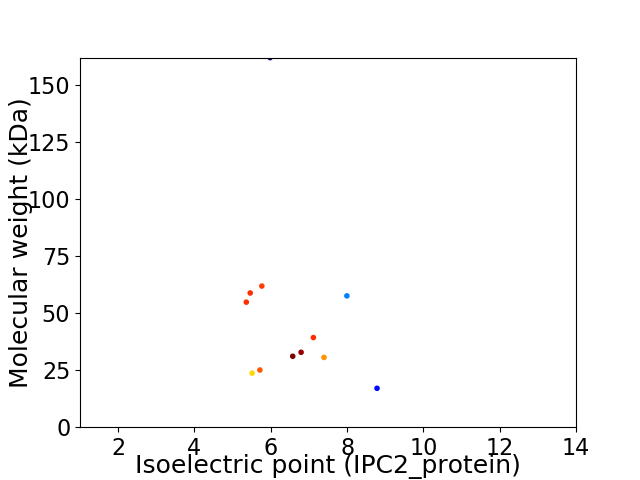
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A499PLS0|A0A499PLS0_9CLOS Uncharacterized protein OS=Pistachio ampelovirus A OX=2093224 PE=4 SV=1
MM1 pKa = 7.56INCFLEE7 pKa = 4.61DD8 pKa = 3.48VVAGSTSLEE17 pKa = 4.02FFHH20 pKa = 8.52RR21 pKa = 11.84EE22 pKa = 3.69DD23 pKa = 3.54CVEE26 pKa = 4.85FEE28 pKa = 5.71DD29 pKa = 6.08FDD31 pKa = 6.28ANLDD35 pKa = 3.62NVVIRR40 pKa = 11.84EE41 pKa = 4.38SEE43 pKa = 4.1NKK45 pKa = 10.27APTRR49 pKa = 11.84NDD51 pKa = 3.19PTTVPVIRR59 pKa = 11.84SQIGSKK65 pKa = 9.99KK66 pKa = 10.32RR67 pKa = 11.84NTLRR71 pKa = 11.84SNLLTFEE78 pKa = 4.33ARR80 pKa = 11.84NFNADD85 pKa = 3.26RR86 pKa = 11.84GVDD89 pKa = 3.72ISCSDD94 pKa = 3.74VVSDD98 pKa = 4.03ILVDD102 pKa = 3.75NFFSRR107 pKa = 11.84CIDD110 pKa = 3.46SSKK113 pKa = 10.97LEE115 pKa = 3.95QANTEE120 pKa = 4.42SINVNLRR127 pKa = 11.84VISDD131 pKa = 3.55WLSSRR136 pKa = 11.84TGTGYY141 pKa = 10.74GALVGEE147 pKa = 4.71LQRR150 pKa = 11.84PSDD153 pKa = 3.88VADD156 pKa = 3.63GLTNFKK162 pKa = 10.75IMVKK166 pKa = 10.18SDD168 pKa = 3.7AKK170 pKa = 10.59PKK172 pKa = 10.06LDD174 pKa = 3.52EE175 pKa = 4.81SIISKK180 pKa = 10.46VSTGQNIVYY189 pKa = 10.07HH190 pKa = 5.84KK191 pKa = 10.39RR192 pKa = 11.84KK193 pKa = 9.79INAIFSNIFLQVVEE207 pKa = 4.3RR208 pKa = 11.84LKK210 pKa = 10.93FALAHH215 pKa = 6.19NIVLYY220 pKa = 10.46TGMNTDD226 pKa = 3.31DD227 pKa = 4.18FARR230 pKa = 11.84EE231 pKa = 3.79VEE233 pKa = 4.36DD234 pKa = 6.49RR235 pKa = 11.84IGCDD239 pKa = 2.67INSFFCSEE247 pKa = 4.25LDD249 pKa = 2.91ISKK252 pKa = 10.56YY253 pKa = 10.61DD254 pKa = 3.49KK255 pKa = 11.33SQGALFKK262 pKa = 10.74QVEE265 pKa = 4.41EE266 pKa = 4.84KK267 pKa = 10.46ILARR271 pKa = 11.84LGVSPGVLDD280 pKa = 3.52LWYY283 pKa = 10.02ASEE286 pKa = 4.47YY287 pKa = 9.6EE288 pKa = 4.37SCVTSADD295 pKa = 3.61RR296 pKa = 11.84TFSGDD301 pKa = 2.14IGAQRR306 pKa = 11.84RR307 pKa = 11.84SGASNTWLGNTLINMCLQSYY327 pKa = 7.96CTDD330 pKa = 3.98FSDD333 pKa = 5.45FPCICFAGDD342 pKa = 3.44DD343 pKa = 3.79SLIFSRR349 pKa = 11.84SPLANVYY356 pKa = 11.04GDD358 pKa = 3.41IQTHH362 pKa = 6.45FGFDD366 pKa = 2.98MKK368 pKa = 10.51FYY370 pKa = 10.44EE371 pKa = 4.05RR372 pKa = 11.84AVPYY376 pKa = 9.95FCSKK380 pKa = 10.92YY381 pKa = 10.39LVSDD385 pKa = 4.17GSKK388 pKa = 10.7LFFVPDD394 pKa = 4.12PYY396 pKa = 11.17KK397 pKa = 11.05VLVKK401 pKa = 10.57LGRR404 pKa = 11.84LTSSKK409 pKa = 10.84EE410 pKa = 4.21SLCHH414 pKa = 6.77EE415 pKa = 4.61NFQSLCDD422 pKa = 3.72SVKK425 pKa = 10.34YY426 pKa = 10.82LNNEE430 pKa = 4.06TIINMLVFYY439 pKa = 7.39HH440 pKa = 6.13HH441 pKa = 7.14CKK443 pKa = 10.48YY444 pKa = 11.02GPTNYY449 pKa = 9.81TYY451 pKa = 11.19GAFASIHH458 pKa = 6.71CITANFSQFRR468 pKa = 11.84RR469 pKa = 11.84LFIRR473 pKa = 11.84RR474 pKa = 11.84NAKK477 pKa = 10.3SFGFDD482 pKa = 4.07LNDD485 pKa = 3.22SS486 pKa = 3.83
MM1 pKa = 7.56INCFLEE7 pKa = 4.61DD8 pKa = 3.48VVAGSTSLEE17 pKa = 4.02FFHH20 pKa = 8.52RR21 pKa = 11.84EE22 pKa = 3.69DD23 pKa = 3.54CVEE26 pKa = 4.85FEE28 pKa = 5.71DD29 pKa = 6.08FDD31 pKa = 6.28ANLDD35 pKa = 3.62NVVIRR40 pKa = 11.84EE41 pKa = 4.38SEE43 pKa = 4.1NKK45 pKa = 10.27APTRR49 pKa = 11.84NDD51 pKa = 3.19PTTVPVIRR59 pKa = 11.84SQIGSKK65 pKa = 9.99KK66 pKa = 10.32RR67 pKa = 11.84NTLRR71 pKa = 11.84SNLLTFEE78 pKa = 4.33ARR80 pKa = 11.84NFNADD85 pKa = 3.26RR86 pKa = 11.84GVDD89 pKa = 3.72ISCSDD94 pKa = 3.74VVSDD98 pKa = 4.03ILVDD102 pKa = 3.75NFFSRR107 pKa = 11.84CIDD110 pKa = 3.46SSKK113 pKa = 10.97LEE115 pKa = 3.95QANTEE120 pKa = 4.42SINVNLRR127 pKa = 11.84VISDD131 pKa = 3.55WLSSRR136 pKa = 11.84TGTGYY141 pKa = 10.74GALVGEE147 pKa = 4.71LQRR150 pKa = 11.84PSDD153 pKa = 3.88VADD156 pKa = 3.63GLTNFKK162 pKa = 10.75IMVKK166 pKa = 10.18SDD168 pKa = 3.7AKK170 pKa = 10.59PKK172 pKa = 10.06LDD174 pKa = 3.52EE175 pKa = 4.81SIISKK180 pKa = 10.46VSTGQNIVYY189 pKa = 10.07HH190 pKa = 5.84KK191 pKa = 10.39RR192 pKa = 11.84KK193 pKa = 9.79INAIFSNIFLQVVEE207 pKa = 4.3RR208 pKa = 11.84LKK210 pKa = 10.93FALAHH215 pKa = 6.19NIVLYY220 pKa = 10.46TGMNTDD226 pKa = 3.31DD227 pKa = 4.18FARR230 pKa = 11.84EE231 pKa = 3.79VEE233 pKa = 4.36DD234 pKa = 6.49RR235 pKa = 11.84IGCDD239 pKa = 2.67INSFFCSEE247 pKa = 4.25LDD249 pKa = 2.91ISKK252 pKa = 10.56YY253 pKa = 10.61DD254 pKa = 3.49KK255 pKa = 11.33SQGALFKK262 pKa = 10.74QVEE265 pKa = 4.41EE266 pKa = 4.84KK267 pKa = 10.46ILARR271 pKa = 11.84LGVSPGVLDD280 pKa = 3.52LWYY283 pKa = 10.02ASEE286 pKa = 4.47YY287 pKa = 9.6EE288 pKa = 4.37SCVTSADD295 pKa = 3.61RR296 pKa = 11.84TFSGDD301 pKa = 2.14IGAQRR306 pKa = 11.84RR307 pKa = 11.84SGASNTWLGNTLINMCLQSYY327 pKa = 7.96CTDD330 pKa = 3.98FSDD333 pKa = 5.45FPCICFAGDD342 pKa = 3.44DD343 pKa = 3.79SLIFSRR349 pKa = 11.84SPLANVYY356 pKa = 11.04GDD358 pKa = 3.41IQTHH362 pKa = 6.45FGFDD366 pKa = 2.98MKK368 pKa = 10.51FYY370 pKa = 10.44EE371 pKa = 4.05RR372 pKa = 11.84AVPYY376 pKa = 9.95FCSKK380 pKa = 10.92YY381 pKa = 10.39LVSDD385 pKa = 4.17GSKK388 pKa = 10.7LFFVPDD394 pKa = 4.12PYY396 pKa = 11.17KK397 pKa = 11.05VLVKK401 pKa = 10.57LGRR404 pKa = 11.84LTSSKK409 pKa = 10.84EE410 pKa = 4.21SLCHH414 pKa = 6.77EE415 pKa = 4.61NFQSLCDD422 pKa = 3.72SVKK425 pKa = 10.34YY426 pKa = 10.82LNNEE430 pKa = 4.06TIINMLVFYY439 pKa = 7.39HH440 pKa = 6.13HH441 pKa = 7.14CKK443 pKa = 10.48YY444 pKa = 11.02GPTNYY449 pKa = 9.81TYY451 pKa = 11.19GAFASIHH458 pKa = 6.71CITANFSQFRR468 pKa = 11.84RR469 pKa = 11.84LFIRR473 pKa = 11.84RR474 pKa = 11.84NAKK477 pKa = 10.3SFGFDD482 pKa = 4.07LNDD485 pKa = 3.22SS486 pKa = 3.83
Molecular weight: 54.83 kDa
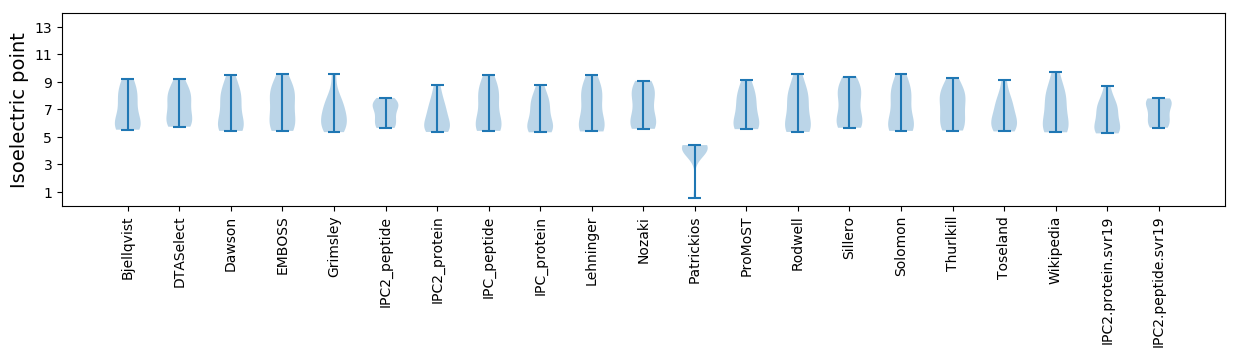
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A499Q1W1|A0A499Q1W1_9CLOS Uncharacterized protein OS=Pistachio ampelovirus A OX=2093224 PE=4 SV=1
MM1 pKa = 7.24SLKK4 pKa = 10.78HH5 pKa = 6.01MGNIISDD12 pKa = 3.76YY13 pKa = 10.54EE14 pKa = 4.0RR15 pKa = 11.84DD16 pKa = 3.69YY17 pKa = 11.94LEE19 pKa = 4.2LNRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GNYY27 pKa = 9.84GEE29 pKa = 3.99PTIVNYY35 pKa = 10.76NGADD39 pKa = 3.3YY40 pKa = 11.11GGYY43 pKa = 9.84RR44 pKa = 11.84RR45 pKa = 11.84LDD47 pKa = 3.09SSNYY51 pKa = 9.33HH52 pKa = 6.12NYY54 pKa = 10.81DD55 pKa = 3.03YY56 pKa = 10.99FDD58 pKa = 3.83PRR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 3.88RR63 pKa = 11.84DD64 pKa = 3.52NKK66 pKa = 10.6LLEE69 pKa = 4.01VWNDD73 pKa = 3.05HH74 pKa = 5.32ATEE77 pKa = 4.53LSQMKK82 pKa = 8.92TRR84 pKa = 11.84EE85 pKa = 3.56TRR87 pKa = 11.84YY88 pKa = 9.36EE89 pKa = 3.6YY90 pKa = 10.35EE91 pKa = 3.68RR92 pKa = 11.84RR93 pKa = 11.84MNIINEE99 pKa = 3.93YY100 pKa = 10.68DD101 pKa = 3.65LEE103 pKa = 4.35LKK105 pKa = 10.24KK106 pKa = 10.74LKK108 pKa = 10.3KK109 pKa = 9.63EE110 pKa = 4.07HH111 pKa = 6.1EE112 pKa = 4.39EE113 pKa = 4.07KK114 pKa = 10.8LKK116 pKa = 10.75KK117 pKa = 10.62AYY119 pKa = 9.48DD120 pKa = 3.2KK121 pKa = 11.03HH122 pKa = 8.05AEE124 pKa = 4.05RR125 pKa = 11.84KK126 pKa = 8.94RR127 pKa = 11.84KK128 pKa = 9.91SFIYY132 pKa = 10.17RR133 pKa = 11.84NLYY136 pKa = 9.17RR137 pKa = 5.6
MM1 pKa = 7.24SLKK4 pKa = 10.78HH5 pKa = 6.01MGNIISDD12 pKa = 3.76YY13 pKa = 10.54EE14 pKa = 4.0RR15 pKa = 11.84DD16 pKa = 3.69YY17 pKa = 11.94LEE19 pKa = 4.2LNRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84GNYY27 pKa = 9.84GEE29 pKa = 3.99PTIVNYY35 pKa = 10.76NGADD39 pKa = 3.3YY40 pKa = 11.11GGYY43 pKa = 9.84RR44 pKa = 11.84RR45 pKa = 11.84LDD47 pKa = 3.09SSNYY51 pKa = 9.33HH52 pKa = 6.12NYY54 pKa = 10.81DD55 pKa = 3.03YY56 pKa = 10.99FDD58 pKa = 3.83PRR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 3.88RR63 pKa = 11.84DD64 pKa = 3.52NKK66 pKa = 10.6LLEE69 pKa = 4.01VWNDD73 pKa = 3.05HH74 pKa = 5.32ATEE77 pKa = 4.53LSQMKK82 pKa = 8.92TRR84 pKa = 11.84EE85 pKa = 3.56TRR87 pKa = 11.84YY88 pKa = 9.36EE89 pKa = 3.6YY90 pKa = 10.35EE91 pKa = 3.68RR92 pKa = 11.84RR93 pKa = 11.84MNIINEE99 pKa = 3.93YY100 pKa = 10.68DD101 pKa = 3.65LEE103 pKa = 4.35LKK105 pKa = 10.24KK106 pKa = 10.74LKK108 pKa = 10.3KK109 pKa = 9.63EE110 pKa = 4.07HH111 pKa = 6.1EE112 pKa = 4.39EE113 pKa = 4.07KK114 pKa = 10.8LKK116 pKa = 10.75KK117 pKa = 10.62AYY119 pKa = 9.48DD120 pKa = 3.2KK121 pKa = 11.03HH122 pKa = 8.05AEE124 pKa = 4.05RR125 pKa = 11.84KK126 pKa = 8.94RR127 pKa = 11.84KK128 pKa = 9.91SFIYY132 pKa = 10.17RR133 pKa = 11.84NLYY136 pKa = 9.17RR137 pKa = 5.6
Molecular weight: 17.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5271 |
137 |
1446 |
439.3 |
49.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.824 ± 0.39 | 1.954 ± 0.407 |
6.109 ± 0.35 | 6.488 ± 0.651 |
4.534 ± 0.463 | 5.559 ± 0.509 |
1.707 ± 0.179 | 6.109 ± 0.588 |
6.242 ± 0.783 | 9.732 ± 0.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.878 ± 0.192 | 5.255 ± 0.391 |
2.903 ± 0.38 | 2.22 ± 0.214 |
5.919 ± 0.34 | 8.329 ± 0.403 |
6.014 ± 0.637 | 8.272 ± 0.546 |
0.74 ± 0.135 | 4.212 ± 0.444 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
