
Coronavirus AcCoV-JC34
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Luchacovirus; unclassified Luchacovirus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
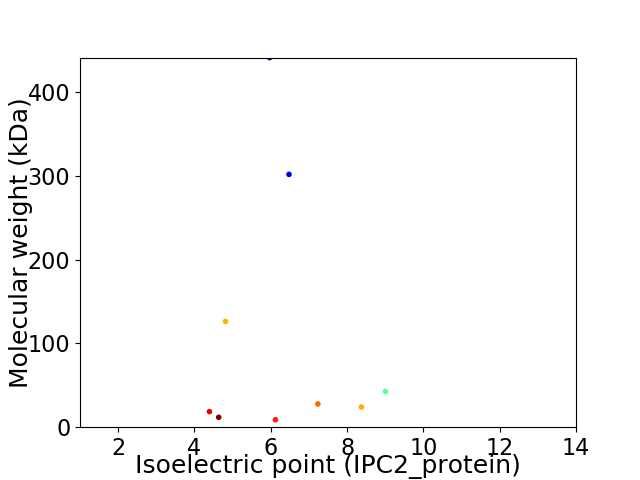
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
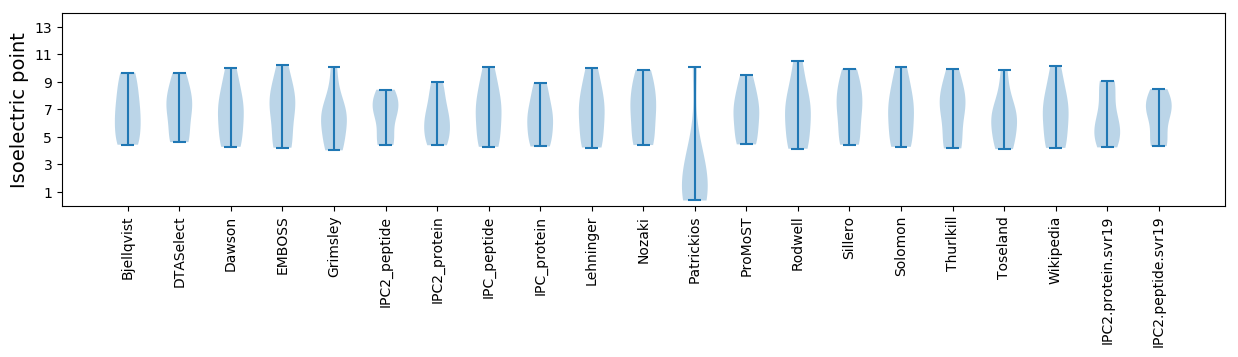
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9JJV5|A0A1X9JJV5_9ALPC Membrane protein OS=Coronavirus AcCoV-JC34 OX=1964806 GN=M PE=3 SV=1
MM1 pKa = 7.47ILVFLVLIASVGAITDD17 pKa = 3.79SKK19 pKa = 10.9SNCSFSFSSLLVDD32 pKa = 3.7GTVSFDD38 pKa = 3.17TVHH41 pKa = 6.85RR42 pKa = 11.84VTFANCFFNWEE53 pKa = 4.1DD54 pKa = 3.3WFGCTSDD61 pKa = 4.53CDD63 pKa = 3.91LMAPLLSAFMPANTSEE79 pKa = 4.05MFQVCNGISVPDD91 pKa = 4.12FDD93 pKa = 4.64NCVSGFVMRR102 pKa = 11.84YY103 pKa = 7.65GALYY107 pKa = 10.62VPDD110 pKa = 4.39DD111 pKa = 3.65THH113 pKa = 7.98VKK115 pKa = 9.8YY116 pKa = 10.94VPEE119 pKa = 4.16KK120 pKa = 10.69NEE122 pKa = 4.2CFGQLKK128 pKa = 10.05FMNNVIWFITSHH140 pKa = 6.22LTNEE144 pKa = 4.31ICCSNRR150 pKa = 11.84IAANPFIPVIEE161 pKa = 4.23NNNFNN166 pKa = 3.92
MM1 pKa = 7.47ILVFLVLIASVGAITDD17 pKa = 3.79SKK19 pKa = 10.9SNCSFSFSSLLVDD32 pKa = 3.7GTVSFDD38 pKa = 3.17TVHH41 pKa = 6.85RR42 pKa = 11.84VTFANCFFNWEE53 pKa = 4.1DD54 pKa = 3.3WFGCTSDD61 pKa = 4.53CDD63 pKa = 3.91LMAPLLSAFMPANTSEE79 pKa = 4.05MFQVCNGISVPDD91 pKa = 4.12FDD93 pKa = 4.64NCVSGFVMRR102 pKa = 11.84YY103 pKa = 7.65GALYY107 pKa = 10.62VPDD110 pKa = 4.39DD111 pKa = 3.65THH113 pKa = 7.98VKK115 pKa = 9.8YY116 pKa = 10.94VPEE119 pKa = 4.16KK120 pKa = 10.69NEE122 pKa = 4.2CFGQLKK128 pKa = 10.05FMNNVIWFITSHH140 pKa = 6.22LTNEE144 pKa = 4.31ICCSNRR150 pKa = 11.84IAANPFIPVIEE161 pKa = 4.23NNNFNN166 pKa = 3.92
Molecular weight: 18.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9JPE2|A0A1X9JPE2_9ALPC Growth factor-like peptide (Fragment) OS=Coronavirus AcCoV-JC34 OX=1964806 PE=3 SV=1
MM1 pKa = 7.57IGGLFSVGFEE11 pKa = 4.7QFIQHH16 pKa = 6.58ANVTTGGALTALAAQPLINYY36 pKa = 6.66GTAVFSVYY44 pKa = 11.03SCFFLSFFALYY55 pKa = 10.1SVKK58 pKa = 10.24SDD60 pKa = 3.5RR61 pKa = 11.84ANCLLLFLRR70 pKa = 11.84LLTLFVAVPILFCTGYY86 pKa = 10.78YY87 pKa = 9.5IDD89 pKa = 5.02GSLTILILLSRR100 pKa = 11.84FCYY103 pKa = 10.45LIYY106 pKa = 10.4YY107 pKa = 7.52CVRR110 pKa = 11.84FKK112 pKa = 10.75RR113 pKa = 11.84LHH115 pKa = 6.25FILYY119 pKa = 7.5NTSTLLFAQGRR130 pKa = 11.84CVPYY134 pKa = 10.92VKK136 pKa = 10.43LHH138 pKa = 5.3YY139 pKa = 9.26FANYY143 pKa = 8.33AALYY147 pKa = 9.7GGAGHH152 pKa = 7.61LMLGRR157 pKa = 11.84KK158 pKa = 8.88VINFTEE164 pKa = 3.89ARR166 pKa = 11.84NVVLAVRR173 pKa = 11.84GRR175 pKa = 11.84LQEE178 pKa = 4.37DD179 pKa = 3.76LLLARR184 pKa = 11.84VVEE187 pKa = 4.3LANGEE192 pKa = 4.46CIYY195 pKa = 10.71IFTKK199 pKa = 10.33EE200 pKa = 4.04PAVSVYY206 pKa = 10.68NFSFQPLNN214 pKa = 3.43
MM1 pKa = 7.57IGGLFSVGFEE11 pKa = 4.7QFIQHH16 pKa = 6.58ANVTTGGALTALAAQPLINYY36 pKa = 6.66GTAVFSVYY44 pKa = 11.03SCFFLSFFALYY55 pKa = 10.1SVKK58 pKa = 10.24SDD60 pKa = 3.5RR61 pKa = 11.84ANCLLLFLRR70 pKa = 11.84LLTLFVAVPILFCTGYY86 pKa = 10.78YY87 pKa = 9.5IDD89 pKa = 5.02GSLTILILLSRR100 pKa = 11.84FCYY103 pKa = 10.45LIYY106 pKa = 10.4YY107 pKa = 7.52CVRR110 pKa = 11.84FKK112 pKa = 10.75RR113 pKa = 11.84LHH115 pKa = 6.25FILYY119 pKa = 7.5NTSTLLFAQGRR130 pKa = 11.84CVPYY134 pKa = 10.92VKK136 pKa = 10.43LHH138 pKa = 5.3YY139 pKa = 9.26FANYY143 pKa = 8.33AALYY147 pKa = 9.7GGAGHH152 pKa = 7.61LMLGRR157 pKa = 11.84KK158 pKa = 8.88VINFTEE164 pKa = 3.89ARR166 pKa = 11.84NVVLAVRR173 pKa = 11.84GRR175 pKa = 11.84LQEE178 pKa = 4.37DD179 pKa = 3.76LLLARR184 pKa = 11.84VVEE187 pKa = 4.3LANGEE192 pKa = 4.46CIYY195 pKa = 10.71IFTKK199 pKa = 10.33EE200 pKa = 4.04PAVSVYY206 pKa = 10.68NFSFQPLNN214 pKa = 3.43
Molecular weight: 24.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8971 |
78 |
3974 |
996.8 |
111.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.755 ± 0.493 | 3.779 ± 0.292 |
5.462 ± 0.336 | 3.712 ± 0.132 |
6.153 ± 0.276 | 6.465 ± 0.247 |
1.995 ± 0.188 | 4.704 ± 0.417 |
5.105 ± 0.625 | 8.84 ± 0.403 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.973 ± 0.247 | 6.12 ± 0.399 |
3.467 ± 0.396 | 2.965 ± 0.453 |
3.478 ± 0.232 | 6.978 ± 0.348 |
5.964 ± 0.286 | 9.676 ± 0.514 |
1.349 ± 0.144 | 5.061 ± 0.39 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
