
Avon-Heathcote Estuary associated circular virus 19
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins
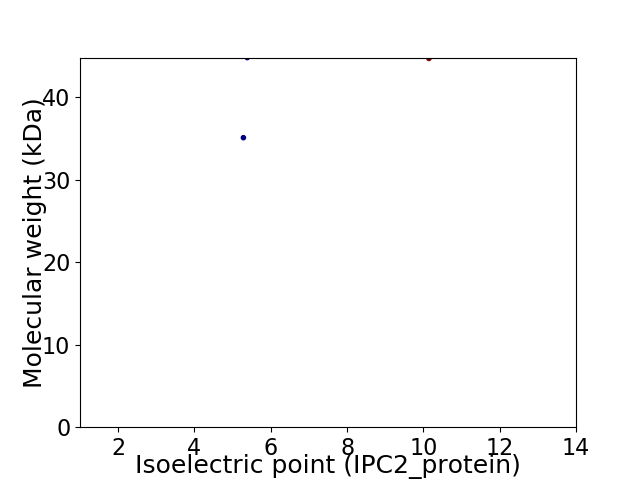
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
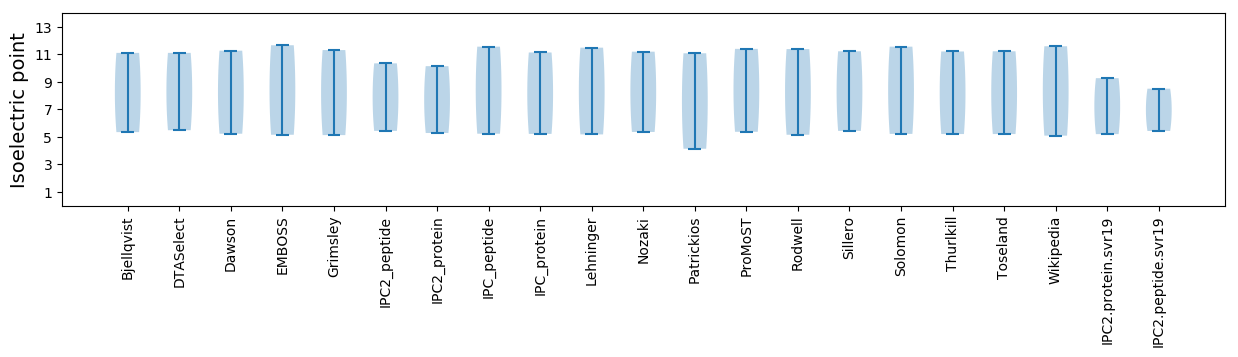
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBR9|A0A0C5IBR9_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 19 OX=1618242 PE=3 SV=1
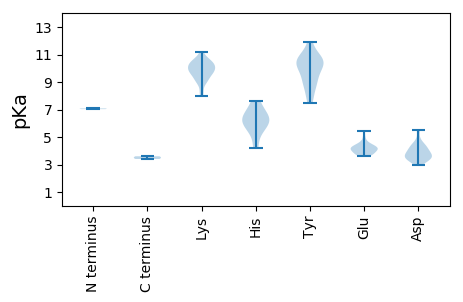
MM1 pKa = 7.03SRR3 pKa = 11.84KK4 pKa = 9.98RR5 pKa = 11.84NWCFTLFPDD14 pKa = 4.06QDD16 pKa = 3.48GYY18 pKa = 10.21YY19 pKa = 9.08TDD21 pKa = 5.42NIADD25 pKa = 4.2MVSGDD30 pKa = 3.91YY31 pKa = 11.03DD32 pKa = 3.75FEE34 pKa = 4.16EE35 pKa = 3.96QKK37 pKa = 10.37IRR39 pKa = 11.84FIIFQEE45 pKa = 4.62EE46 pKa = 4.2ICPEE50 pKa = 3.69SSRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.23LQGYY59 pKa = 7.96VEE61 pKa = 4.36YY62 pKa = 9.52TVSRR66 pKa = 11.84RR67 pKa = 11.84LAAVKK72 pKa = 10.33LSLGSEE78 pKa = 4.41SVHH81 pKa = 7.63LEE83 pKa = 3.6GRR85 pKa = 11.84RR86 pKa = 11.84GTASEE91 pKa = 3.7AAEE94 pKa = 4.12YY95 pKa = 10.55CEE97 pKa = 5.47KK98 pKa = 10.49EE99 pKa = 4.75DD100 pKa = 4.4SFAPDD105 pKa = 4.37GIRR108 pKa = 11.84FQWGEE113 pKa = 3.76LGGEE117 pKa = 3.79QGARR121 pKa = 11.84SDD123 pKa = 3.84LATVASAVISGKK135 pKa = 9.92RR136 pKa = 11.84VEE138 pKa = 4.4EE139 pKa = 4.13VALEE143 pKa = 4.14FPTTFIRR150 pKa = 11.84YY151 pKa = 8.54NRR153 pKa = 11.84GIEE156 pKa = 3.86KK157 pKa = 8.96LTNVVRR163 pKa = 11.84RR164 pKa = 11.84KK165 pKa = 9.05EE166 pKa = 3.79QMRR169 pKa = 11.84NLRR172 pKa = 11.84MLKK175 pKa = 10.26VVVLWGDD182 pKa = 3.03AGTGKK187 pKa = 8.25TRR189 pKa = 11.84TAFEE193 pKa = 5.05LSPDD197 pKa = 3.65HH198 pKa = 7.06YY199 pKa = 10.92ILSQQDD205 pKa = 3.29GQLWWDD211 pKa = 3.86GYY213 pKa = 10.88EE214 pKa = 4.32GQKK217 pKa = 10.61HH218 pKa = 6.5LIIDD222 pKa = 4.68DD223 pKa = 4.37FYY225 pKa = 11.9GWIKK229 pKa = 9.98WGLFLRR235 pKa = 11.84ILDD238 pKa = 4.5IYY240 pKa = 10.24PLQLPVKK247 pKa = 9.71GGFTIASWDD256 pKa = 3.59VVFITSNCMPRR267 pKa = 11.84NWYY270 pKa = 8.71EE271 pKa = 3.75RR272 pKa = 11.84GMPAEE277 pKa = 4.1LARR280 pKa = 11.84RR281 pKa = 11.84LTHH284 pKa = 6.26IVEE287 pKa = 4.37FEE289 pKa = 4.1SYY291 pKa = 8.62EE292 pKa = 3.99TDD294 pKa = 3.45FDD296 pKa = 4.5KK297 pKa = 11.02IKK299 pKa = 10.66EE300 pKa = 4.08ILVV303 pKa = 3.6
MM1 pKa = 7.03SRR3 pKa = 11.84KK4 pKa = 9.98RR5 pKa = 11.84NWCFTLFPDD14 pKa = 4.06QDD16 pKa = 3.48GYY18 pKa = 10.21YY19 pKa = 9.08TDD21 pKa = 5.42NIADD25 pKa = 4.2MVSGDD30 pKa = 3.91YY31 pKa = 11.03DD32 pKa = 3.75FEE34 pKa = 4.16EE35 pKa = 3.96QKK37 pKa = 10.37IRR39 pKa = 11.84FIIFQEE45 pKa = 4.62EE46 pKa = 4.2ICPEE50 pKa = 3.69SSRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.23LQGYY59 pKa = 7.96VEE61 pKa = 4.36YY62 pKa = 9.52TVSRR66 pKa = 11.84RR67 pKa = 11.84LAAVKK72 pKa = 10.33LSLGSEE78 pKa = 4.41SVHH81 pKa = 7.63LEE83 pKa = 3.6GRR85 pKa = 11.84RR86 pKa = 11.84GTASEE91 pKa = 3.7AAEE94 pKa = 4.12YY95 pKa = 10.55CEE97 pKa = 5.47KK98 pKa = 10.49EE99 pKa = 4.75DD100 pKa = 4.4SFAPDD105 pKa = 4.37GIRR108 pKa = 11.84FQWGEE113 pKa = 3.76LGGEE117 pKa = 3.79QGARR121 pKa = 11.84SDD123 pKa = 3.84LATVASAVISGKK135 pKa = 9.92RR136 pKa = 11.84VEE138 pKa = 4.4EE139 pKa = 4.13VALEE143 pKa = 4.14FPTTFIRR150 pKa = 11.84YY151 pKa = 8.54NRR153 pKa = 11.84GIEE156 pKa = 3.86KK157 pKa = 8.96LTNVVRR163 pKa = 11.84RR164 pKa = 11.84KK165 pKa = 9.05EE166 pKa = 3.79QMRR169 pKa = 11.84NLRR172 pKa = 11.84MLKK175 pKa = 10.26VVVLWGDD182 pKa = 3.03AGTGKK187 pKa = 8.25TRR189 pKa = 11.84TAFEE193 pKa = 5.05LSPDD197 pKa = 3.65HH198 pKa = 7.06YY199 pKa = 10.92ILSQQDD205 pKa = 3.29GQLWWDD211 pKa = 3.86GYY213 pKa = 10.88EE214 pKa = 4.32GQKK217 pKa = 10.61HH218 pKa = 6.5LIIDD222 pKa = 4.68DD223 pKa = 4.37FYY225 pKa = 11.9GWIKK229 pKa = 9.98WGLFLRR235 pKa = 11.84ILDD238 pKa = 4.5IYY240 pKa = 10.24PLQLPVKK247 pKa = 9.71GGFTIASWDD256 pKa = 3.59VVFITSNCMPRR267 pKa = 11.84NWYY270 pKa = 8.71EE271 pKa = 3.75RR272 pKa = 11.84GMPAEE277 pKa = 4.1LARR280 pKa = 11.84RR281 pKa = 11.84LTHH284 pKa = 6.26IVEE287 pKa = 4.37FEE289 pKa = 4.1SYY291 pKa = 8.62EE292 pKa = 3.99TDD294 pKa = 3.45FDD296 pKa = 4.5KK297 pKa = 11.02IKK299 pKa = 10.66EE300 pKa = 4.08ILVV303 pKa = 3.6
Molecular weight: 35.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBR9|A0A0C5IBR9_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 19 OX=1618242 PE=3 SV=1
MM1 pKa = 7.08ARR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.16KK9 pKa = 10.35KK10 pKa = 9.62SSRR13 pKa = 11.84SRR15 pKa = 11.84WSFPALHH22 pKa = 6.02YY23 pKa = 10.29PGYY26 pKa = 10.54NYY28 pKa = 10.52LGPGTKK34 pKa = 10.32NFDD37 pKa = 3.38RR38 pKa = 11.84APRR41 pKa = 11.84NDD43 pKa = 3.53LDD45 pKa = 3.69AAARR49 pKa = 11.84KK50 pKa = 9.45HH51 pKa = 6.2DD52 pKa = 4.19LDD54 pKa = 3.69YY55 pKa = 11.77GKK57 pKa = 10.12LQRR60 pKa = 11.84GGRR63 pKa = 11.84NPYY66 pKa = 9.92FKK68 pKa = 10.08WFKK71 pKa = 10.59ADD73 pKa = 3.38QRR75 pKa = 11.84FLDD78 pKa = 5.5RR79 pKa = 11.84IAKK82 pKa = 9.77DD83 pKa = 3.18KK84 pKa = 11.19SVPGWIARR92 pKa = 11.84QTFRR96 pKa = 11.84AKK98 pKa = 10.36KK99 pKa = 8.98YY100 pKa = 7.76FAPQGDD106 pKa = 4.25EE107 pKa = 4.33LPRR110 pKa = 11.84KK111 pKa = 8.29HH112 pKa = 5.69TGKK115 pKa = 10.24RR116 pKa = 11.84SRR118 pKa = 11.84SAIPVYY124 pKa = 10.68LIDD127 pKa = 4.14RR128 pKa = 11.84FSHH131 pKa = 6.55KK132 pKa = 9.96KK133 pKa = 9.74QKK135 pKa = 9.27TLGGSLKK142 pKa = 10.46KK143 pKa = 9.76MRR145 pKa = 11.84RR146 pKa = 11.84TWRR149 pKa = 11.84RR150 pKa = 11.84PRR152 pKa = 11.84ALKK155 pKa = 10.14RR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84SASYY162 pKa = 9.96GRR164 pKa = 11.84KK165 pKa = 8.91RR166 pKa = 11.84RR167 pKa = 11.84YY168 pKa = 9.07KK169 pKa = 9.14RR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84YY173 pKa = 9.83PSMKK177 pKa = 9.59PLYY180 pKa = 10.05RR181 pKa = 11.84KK182 pKa = 9.53IKK184 pKa = 10.15RR185 pKa = 11.84VAKK188 pKa = 10.02GLGRR192 pKa = 11.84MQHH195 pKa = 5.4QNAVANVYY203 pKa = 10.53RR204 pKa = 11.84DD205 pKa = 3.59VSSGNVSWDD214 pKa = 3.33AGEE217 pKa = 4.47CAYY220 pKa = 10.18KK221 pKa = 10.61DD222 pKa = 2.94ITAILTSGQMDD233 pKa = 3.51NVLSTFRR240 pKa = 11.84VLQMDD245 pKa = 3.82AGVNSRR251 pKa = 11.84EE252 pKa = 4.11TLDD255 pKa = 4.88LDD257 pKa = 4.07TGTATANVKK266 pKa = 9.55FVNARR271 pKa = 11.84SRR273 pKa = 11.84TVFRR277 pKa = 11.84NNTTQRR283 pKa = 11.84ANIAVYY289 pKa = 10.74VLTPTQDD296 pKa = 3.35LSNTPSSLVSSGLTTAEE313 pKa = 3.8ATAPSIDD320 pKa = 3.23IKK322 pKa = 10.64YY323 pKa = 9.27WPVDD327 pKa = 3.45SPIFRR332 pKa = 11.84RR333 pKa = 11.84YY334 pKa = 7.48WKK336 pKa = 9.81IVKK339 pKa = 9.83KK340 pKa = 9.2RR341 pKa = 11.84TRR343 pKa = 11.84ALDD346 pKa = 4.01PGDD349 pKa = 3.86EE350 pKa = 4.33MTVMMRR356 pKa = 11.84RR357 pKa = 11.84NKK359 pKa = 9.7PFSWNNKK366 pKa = 7.99PNDD369 pKa = 3.66VNAEE373 pKa = 4.13SYY375 pKa = 10.88RR376 pKa = 11.84GMKK379 pKa = 9.22SQVVMVRR386 pKa = 3.41
MM1 pKa = 7.08ARR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.16KK9 pKa = 10.35KK10 pKa = 9.62SSRR13 pKa = 11.84SRR15 pKa = 11.84WSFPALHH22 pKa = 6.02YY23 pKa = 10.29PGYY26 pKa = 10.54NYY28 pKa = 10.52LGPGTKK34 pKa = 10.32NFDD37 pKa = 3.38RR38 pKa = 11.84APRR41 pKa = 11.84NDD43 pKa = 3.53LDD45 pKa = 3.69AAARR49 pKa = 11.84KK50 pKa = 9.45HH51 pKa = 6.2DD52 pKa = 4.19LDD54 pKa = 3.69YY55 pKa = 11.77GKK57 pKa = 10.12LQRR60 pKa = 11.84GGRR63 pKa = 11.84NPYY66 pKa = 9.92FKK68 pKa = 10.08WFKK71 pKa = 10.59ADD73 pKa = 3.38QRR75 pKa = 11.84FLDD78 pKa = 5.5RR79 pKa = 11.84IAKK82 pKa = 9.77DD83 pKa = 3.18KK84 pKa = 11.19SVPGWIARR92 pKa = 11.84QTFRR96 pKa = 11.84AKK98 pKa = 10.36KK99 pKa = 8.98YY100 pKa = 7.76FAPQGDD106 pKa = 4.25EE107 pKa = 4.33LPRR110 pKa = 11.84KK111 pKa = 8.29HH112 pKa = 5.69TGKK115 pKa = 10.24RR116 pKa = 11.84SRR118 pKa = 11.84SAIPVYY124 pKa = 10.68LIDD127 pKa = 4.14RR128 pKa = 11.84FSHH131 pKa = 6.55KK132 pKa = 9.96KK133 pKa = 9.74QKK135 pKa = 9.27TLGGSLKK142 pKa = 10.46KK143 pKa = 9.76MRR145 pKa = 11.84RR146 pKa = 11.84TWRR149 pKa = 11.84RR150 pKa = 11.84PRR152 pKa = 11.84ALKK155 pKa = 10.14RR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84SASYY162 pKa = 9.96GRR164 pKa = 11.84KK165 pKa = 8.91RR166 pKa = 11.84RR167 pKa = 11.84YY168 pKa = 9.07KK169 pKa = 9.14RR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84YY173 pKa = 9.83PSMKK177 pKa = 9.59PLYY180 pKa = 10.05RR181 pKa = 11.84KK182 pKa = 9.53IKK184 pKa = 10.15RR185 pKa = 11.84VAKK188 pKa = 10.02GLGRR192 pKa = 11.84MQHH195 pKa = 5.4QNAVANVYY203 pKa = 10.53RR204 pKa = 11.84DD205 pKa = 3.59VSSGNVSWDD214 pKa = 3.33AGEE217 pKa = 4.47CAYY220 pKa = 10.18KK221 pKa = 10.61DD222 pKa = 2.94ITAILTSGQMDD233 pKa = 3.51NVLSTFRR240 pKa = 11.84VLQMDD245 pKa = 3.82AGVNSRR251 pKa = 11.84EE252 pKa = 4.11TLDD255 pKa = 4.88LDD257 pKa = 4.07TGTATANVKK266 pKa = 9.55FVNARR271 pKa = 11.84SRR273 pKa = 11.84TVFRR277 pKa = 11.84NNTTQRR283 pKa = 11.84ANIAVYY289 pKa = 10.74VLTPTQDD296 pKa = 3.35LSNTPSSLVSSGLTTAEE313 pKa = 3.8ATAPSIDD320 pKa = 3.23IKK322 pKa = 10.64YY323 pKa = 9.27WPVDD327 pKa = 3.45SPIFRR332 pKa = 11.84RR333 pKa = 11.84YY334 pKa = 7.48WKK336 pKa = 9.81IVKK339 pKa = 9.83KK340 pKa = 9.2RR341 pKa = 11.84TRR343 pKa = 11.84ALDD346 pKa = 4.01PGDD349 pKa = 3.86EE350 pKa = 4.33MTVMMRR356 pKa = 11.84RR357 pKa = 11.84NKK359 pKa = 9.7PFSWNNKK366 pKa = 7.99PNDD369 pKa = 3.66VNAEE373 pKa = 4.13SYY375 pKa = 10.88RR376 pKa = 11.84GMKK379 pKa = 9.22SQVVMVRR386 pKa = 3.41
Molecular weight: 44.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
689 |
303 |
386 |
344.5 |
39.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.967 ± 0.861 | 0.726 ± 0.377 |
6.096 ± 0.111 | 4.79 ± 2.616 |
4.209 ± 0.68 | 6.676 ± 0.79 |
1.451 ± 0.126 | 4.644 ± 1.241 |
6.967 ± 1.489 | 6.967 ± 0.815 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.309 | 3.919 ± 1.021 |
4.209 ± 0.786 | 3.483 ± 0.303 |
10.885 ± 1.881 | 6.967 ± 0.651 |
5.515 ± 0.358 | 6.096 ± 0.111 |
2.467 ± 0.319 | 4.499 ± 0.133 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
