
Ixcanal virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Ixcanal phlebovirus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
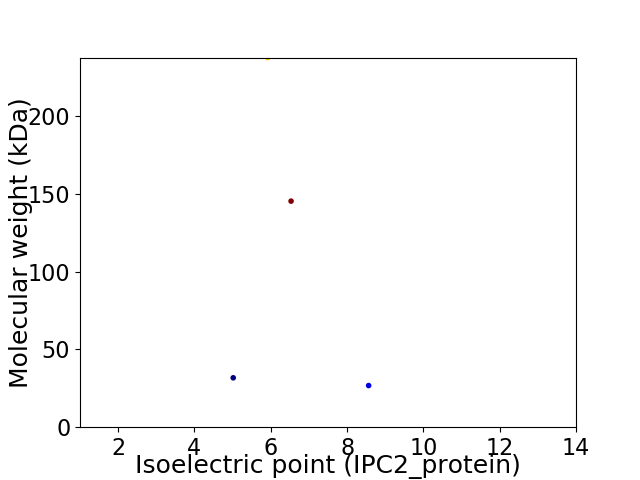
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4ZCL3|F4ZCL3_9VIRU Nucleoprotein OS=Ixcanal virus OX=1006586 GN=NP PE=3 SV=1
MM1 pKa = 7.42TSYY4 pKa = 11.29LFDD7 pKa = 3.42TPVIVRR13 pKa = 11.84PNGYY17 pKa = 9.24FDD19 pKa = 4.06RR20 pKa = 11.84EE21 pKa = 4.37VYY23 pKa = 7.9VTYY26 pKa = 9.66MAHH29 pKa = 6.83NKK31 pKa = 9.73YY32 pKa = 9.65CDD34 pKa = 3.61HH35 pKa = 7.05PVSVHH40 pKa = 5.37NCMEE44 pKa = 4.16IPVKK48 pKa = 10.93NFGVSLLHH56 pKa = 6.9KK57 pKa = 10.19EE58 pKa = 4.29SLSDD62 pKa = 4.34FYY64 pKa = 11.16ILGLLPFRR72 pKa = 11.84WGDD75 pKa = 3.04KK76 pKa = 9.48MGNSRR81 pKa = 11.84VTRR84 pKa = 11.84DD85 pKa = 3.28SFPFMRR91 pKa = 11.84DD92 pKa = 2.97LLNDD96 pKa = 3.42INDD99 pKa = 3.52MHH101 pKa = 8.0DD102 pKa = 3.48SEE104 pKa = 5.35FFISFLPNVKK114 pKa = 9.99KK115 pKa = 10.69ALSWPLGYY123 pKa = 7.63PTLDD127 pKa = 4.63FIRR130 pKa = 11.84MTRR133 pKa = 11.84SEE135 pKa = 4.11LDD137 pKa = 2.93IPHH140 pKa = 6.26FRR142 pKa = 11.84EE143 pKa = 3.72ISSNATLIMQMGQPAIDD160 pKa = 4.12LDD162 pKa = 3.78QGFVNAHH169 pKa = 5.65KK170 pKa = 10.07RR171 pKa = 11.84IVMEE175 pKa = 3.94SVTRR179 pKa = 11.84GFGHH183 pKa = 7.39KK184 pKa = 10.24DD185 pKa = 3.35FPGKK189 pKa = 10.59NMIFEE194 pKa = 4.4VASLQCVRR202 pKa = 11.84LLNAVPADD210 pKa = 3.79LLYY213 pKa = 11.13CQVTNSLTSTVMKK226 pKa = 9.65HH227 pKa = 5.96RR228 pKa = 11.84SMLEE232 pKa = 3.66DD233 pKa = 4.0HH234 pKa = 6.88PQHH237 pKa = 6.87PLGNQKK243 pKa = 7.49WTPDD247 pKa = 3.73PDD249 pKa = 3.18SWLLDD254 pKa = 3.61DD255 pKa = 5.13QAATLHH261 pKa = 6.66EE262 pKa = 4.94DD263 pKa = 3.26FALILSEE270 pKa = 5.18SDD272 pKa = 3.71DD273 pKa = 5.56DD274 pKa = 4.45EE275 pKa = 5.01DD276 pKa = 4.25
MM1 pKa = 7.42TSYY4 pKa = 11.29LFDD7 pKa = 3.42TPVIVRR13 pKa = 11.84PNGYY17 pKa = 9.24FDD19 pKa = 4.06RR20 pKa = 11.84EE21 pKa = 4.37VYY23 pKa = 7.9VTYY26 pKa = 9.66MAHH29 pKa = 6.83NKK31 pKa = 9.73YY32 pKa = 9.65CDD34 pKa = 3.61HH35 pKa = 7.05PVSVHH40 pKa = 5.37NCMEE44 pKa = 4.16IPVKK48 pKa = 10.93NFGVSLLHH56 pKa = 6.9KK57 pKa = 10.19EE58 pKa = 4.29SLSDD62 pKa = 4.34FYY64 pKa = 11.16ILGLLPFRR72 pKa = 11.84WGDD75 pKa = 3.04KK76 pKa = 9.48MGNSRR81 pKa = 11.84VTRR84 pKa = 11.84DD85 pKa = 3.28SFPFMRR91 pKa = 11.84DD92 pKa = 2.97LLNDD96 pKa = 3.42INDD99 pKa = 3.52MHH101 pKa = 8.0DD102 pKa = 3.48SEE104 pKa = 5.35FFISFLPNVKK114 pKa = 9.99KK115 pKa = 10.69ALSWPLGYY123 pKa = 7.63PTLDD127 pKa = 4.63FIRR130 pKa = 11.84MTRR133 pKa = 11.84SEE135 pKa = 4.11LDD137 pKa = 2.93IPHH140 pKa = 6.26FRR142 pKa = 11.84EE143 pKa = 3.72ISSNATLIMQMGQPAIDD160 pKa = 4.12LDD162 pKa = 3.78QGFVNAHH169 pKa = 5.65KK170 pKa = 10.07RR171 pKa = 11.84IVMEE175 pKa = 3.94SVTRR179 pKa = 11.84GFGHH183 pKa = 7.39KK184 pKa = 10.24DD185 pKa = 3.35FPGKK189 pKa = 10.59NMIFEE194 pKa = 4.4VASLQCVRR202 pKa = 11.84LLNAVPADD210 pKa = 3.79LLYY213 pKa = 11.13CQVTNSLTSTVMKK226 pKa = 9.65HH227 pKa = 5.96RR228 pKa = 11.84SMLEE232 pKa = 3.66DD233 pKa = 4.0HH234 pKa = 6.88PQHH237 pKa = 6.87PLGNQKK243 pKa = 7.49WTPDD247 pKa = 3.73PDD249 pKa = 3.18SWLLDD254 pKa = 3.61DD255 pKa = 5.13QAATLHH261 pKa = 6.66EE262 pKa = 4.94DD263 pKa = 3.26FALILSEE270 pKa = 5.18SDD272 pKa = 3.71DD273 pKa = 5.56DD274 pKa = 4.45EE275 pKa = 5.01DD276 pKa = 4.25
Molecular weight: 31.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4ZCL4|F4ZCL4_9VIRU Replicase OS=Ixcanal virus OX=1006586 GN=L PE=4 SV=1
MM1 pKa = 7.94ADD3 pKa = 3.97FEE5 pKa = 4.33RR6 pKa = 11.84LAIEE10 pKa = 4.29FSEE13 pKa = 4.5AGVNIADD20 pKa = 3.57VVNWVNEE27 pKa = 3.75FAYY30 pKa = 10.45QGFDD34 pKa = 2.96AKK36 pKa = 10.46RR37 pKa = 11.84VLEE40 pKa = 4.07LLQQRR45 pKa = 11.84GGSSWQEE52 pKa = 4.09DD53 pKa = 3.61ARR55 pKa = 11.84KK56 pKa = 9.14MIVLALTRR64 pKa = 11.84GNKK67 pKa = 8.15IEE69 pKa = 4.3KK70 pKa = 8.97MVLKK74 pKa = 10.19MSEE77 pKa = 4.1EE78 pKa = 4.2GKK80 pKa = 8.63KK81 pKa = 8.61TVLALKK87 pKa = 10.23KK88 pKa = 10.2KK89 pKa = 9.41YY90 pKa = 7.98QLKK93 pKa = 10.32SGNPGRR99 pKa = 11.84DD100 pKa = 3.45DD101 pKa = 3.54LTLSRR106 pKa = 11.84VAAALAGWTCQALPHH121 pKa = 6.37VEE123 pKa = 3.94NFLPVTGEE131 pKa = 3.97AMDD134 pKa = 4.37ALSPGFPRR142 pKa = 11.84CLMHH146 pKa = 7.07PSFAGLVDD154 pKa = 3.56TTLPPDD160 pKa = 3.66TQDD163 pKa = 4.43AILAAHH169 pKa = 6.45SLFLVQFSKK178 pKa = 11.17VINPNLRR185 pKa = 11.84GKK187 pKa = 9.78PKK189 pKa = 10.67GEE191 pKa = 3.8VVASFKK197 pKa = 11.0QPMLAAVNGGFIGPDD212 pKa = 2.71KK213 pKa = 10.31RR214 pKa = 11.84RR215 pKa = 11.84RR216 pKa = 11.84MLQSLGVVDD225 pKa = 4.2VNGVPSEE232 pKa = 4.39AIKK235 pKa = 10.7VAANKK240 pKa = 9.18FRR242 pKa = 11.84NSQQ245 pKa = 3.01
MM1 pKa = 7.94ADD3 pKa = 3.97FEE5 pKa = 4.33RR6 pKa = 11.84LAIEE10 pKa = 4.29FSEE13 pKa = 4.5AGVNIADD20 pKa = 3.57VVNWVNEE27 pKa = 3.75FAYY30 pKa = 10.45QGFDD34 pKa = 2.96AKK36 pKa = 10.46RR37 pKa = 11.84VLEE40 pKa = 4.07LLQQRR45 pKa = 11.84GGSSWQEE52 pKa = 4.09DD53 pKa = 3.61ARR55 pKa = 11.84KK56 pKa = 9.14MIVLALTRR64 pKa = 11.84GNKK67 pKa = 8.15IEE69 pKa = 4.3KK70 pKa = 8.97MVLKK74 pKa = 10.19MSEE77 pKa = 4.1EE78 pKa = 4.2GKK80 pKa = 8.63KK81 pKa = 8.61TVLALKK87 pKa = 10.23KK88 pKa = 10.2KK89 pKa = 9.41YY90 pKa = 7.98QLKK93 pKa = 10.32SGNPGRR99 pKa = 11.84DD100 pKa = 3.45DD101 pKa = 3.54LTLSRR106 pKa = 11.84VAAALAGWTCQALPHH121 pKa = 6.37VEE123 pKa = 3.94NFLPVTGEE131 pKa = 3.97AMDD134 pKa = 4.37ALSPGFPRR142 pKa = 11.84CLMHH146 pKa = 7.07PSFAGLVDD154 pKa = 3.56TTLPPDD160 pKa = 3.66TQDD163 pKa = 4.43AILAAHH169 pKa = 6.45SLFLVQFSKK178 pKa = 11.17VINPNLRR185 pKa = 11.84GKK187 pKa = 9.78PKK189 pKa = 10.67GEE191 pKa = 3.8VVASFKK197 pKa = 11.0QPMLAAVNGGFIGPDD212 pKa = 2.71KK213 pKa = 10.31RR214 pKa = 11.84RR215 pKa = 11.84RR216 pKa = 11.84MLQSLGVVDD225 pKa = 4.2VNGVPSEE232 pKa = 4.39AIKK235 pKa = 10.7VAANKK240 pKa = 9.18FRR242 pKa = 11.84NSQQ245 pKa = 3.01
Molecular weight: 26.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3915 |
245 |
2085 |
978.8 |
110.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.879 ± 0.822 | 2.81 ± 0.976 |
5.875 ± 0.386 | 6.743 ± 0.68 |
4.623 ± 0.199 | 6.156 ± 0.287 |
2.452 ± 0.195 | 6.079 ± 0.393 |
6.309 ± 0.334 | 9.119 ± 0.622 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.116 ± 0.271 | 3.704 ± 0.267 |
3.959 ± 0.339 | 3.193 ± 0.166 |
5.594 ± 0.26 | 9.834 ± 0.592 |
4.904 ± 0.286 | 6.692 ± 0.235 |
1.303 ± 0.114 | 2.656 ± 0.185 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
