
Agrobacterium phage Atu_ph04
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Ackermannviridae; unclassified Ackermannviridae
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
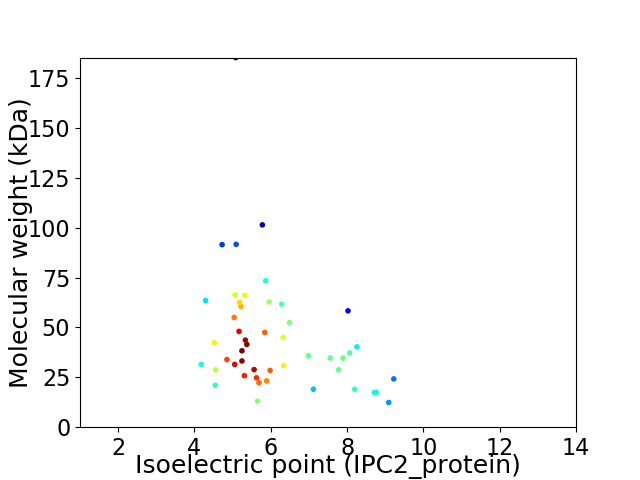
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
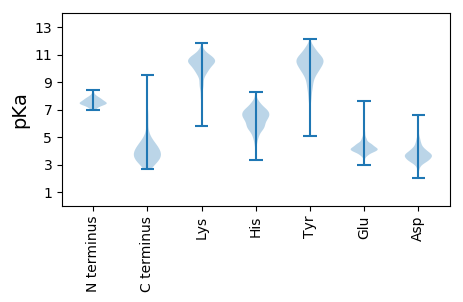
>tr|A0A223VZU8|A0A223VZU8_9CAUD Topoisomerase IV subunit A OS=Agrobacterium phage Atu_ph04 OX=2024263 PE=4 SV=2
MM1 pKa = 7.19VNPYY5 pKa = 9.87FKK7 pKa = 11.21SNFFDD12 pKa = 4.18NEE14 pKa = 4.03MNLLADD20 pKa = 4.15LTDD23 pKa = 3.42EE24 pKa = 5.44AIYY27 pKa = 8.11TTGIEE32 pKa = 4.22VEE34 pKa = 4.18YY35 pKa = 10.33LPRR38 pKa = 11.84KK39 pKa = 9.1IVQHH43 pKa = 6.17DD44 pKa = 4.18PIMNEE49 pKa = 3.82PLSSQFNEE57 pKa = 4.63VYY59 pKa = 10.35QIDD62 pKa = 3.86MAIEE66 pKa = 4.04STEE69 pKa = 4.04SFFAGNEE76 pKa = 4.32SISSIGLAFSFASSSLSVSRR96 pKa = 11.84RR97 pKa = 11.84KK98 pKa = 8.47FTSVTGMQEE107 pKa = 3.98PLNGDD112 pKa = 4.42LIFIKK117 pKa = 9.97QNQMLFEE124 pKa = 4.18IVNTNVRR131 pKa = 11.84DD132 pKa = 3.72PLISGGRR139 pKa = 11.84HH140 pKa = 3.34FTFTIFVKK148 pKa = 10.22PFSIGEE154 pKa = 4.26GNSSFKK160 pKa = 10.39EE161 pKa = 4.25SKK163 pKa = 10.6FKK165 pKa = 10.85GSRR168 pKa = 11.84DD169 pKa = 3.57LQEE172 pKa = 4.12TLSNFLGVVDD182 pKa = 3.84QKK184 pKa = 11.47EE185 pKa = 4.11WDD187 pKa = 3.85FLDD190 pKa = 4.16ASVDD194 pKa = 4.2SVTADD199 pKa = 3.41NNVLRR204 pKa = 11.84VDD206 pKa = 3.81VEE208 pKa = 4.26EE209 pKa = 4.99SGLSQIVRR217 pKa = 11.84ADD219 pKa = 3.65SEE221 pKa = 4.88TFTADD226 pKa = 4.05CEE228 pKa = 4.35LTADD232 pKa = 4.62DD233 pKa = 6.62LLGVWNNSIDD243 pKa = 3.91DD244 pKa = 3.74VMRR247 pKa = 11.84TMDD250 pKa = 3.32QKK252 pKa = 11.09IPNFSDD258 pKa = 2.95NGEE261 pKa = 4.19FEE263 pKa = 4.41CVEE266 pKa = 4.08EE267 pKa = 5.34KK268 pKa = 10.82ILVDD272 pKa = 3.15KK273 pKa = 9.67TNPFGFFF280 pKa = 3.32
MM1 pKa = 7.19VNPYY5 pKa = 9.87FKK7 pKa = 11.21SNFFDD12 pKa = 4.18NEE14 pKa = 4.03MNLLADD20 pKa = 4.15LTDD23 pKa = 3.42EE24 pKa = 5.44AIYY27 pKa = 8.11TTGIEE32 pKa = 4.22VEE34 pKa = 4.18YY35 pKa = 10.33LPRR38 pKa = 11.84KK39 pKa = 9.1IVQHH43 pKa = 6.17DD44 pKa = 4.18PIMNEE49 pKa = 3.82PLSSQFNEE57 pKa = 4.63VYY59 pKa = 10.35QIDD62 pKa = 3.86MAIEE66 pKa = 4.04STEE69 pKa = 4.04SFFAGNEE76 pKa = 4.32SISSIGLAFSFASSSLSVSRR96 pKa = 11.84RR97 pKa = 11.84KK98 pKa = 8.47FTSVTGMQEE107 pKa = 3.98PLNGDD112 pKa = 4.42LIFIKK117 pKa = 9.97QNQMLFEE124 pKa = 4.18IVNTNVRR131 pKa = 11.84DD132 pKa = 3.72PLISGGRR139 pKa = 11.84HH140 pKa = 3.34FTFTIFVKK148 pKa = 10.22PFSIGEE154 pKa = 4.26GNSSFKK160 pKa = 10.39EE161 pKa = 4.25SKK163 pKa = 10.6FKK165 pKa = 10.85GSRR168 pKa = 11.84DD169 pKa = 3.57LQEE172 pKa = 4.12TLSNFLGVVDD182 pKa = 3.84QKK184 pKa = 11.47EE185 pKa = 4.11WDD187 pKa = 3.85FLDD190 pKa = 4.16ASVDD194 pKa = 4.2SVTADD199 pKa = 3.41NNVLRR204 pKa = 11.84VDD206 pKa = 3.81VEE208 pKa = 4.26EE209 pKa = 4.99SGLSQIVRR217 pKa = 11.84ADD219 pKa = 3.65SEE221 pKa = 4.88TFTADD226 pKa = 4.05CEE228 pKa = 4.35LTADD232 pKa = 4.62DD233 pKa = 6.62LLGVWNNSIDD243 pKa = 3.91DD244 pKa = 3.74VMRR247 pKa = 11.84TMDD250 pKa = 3.32QKK252 pKa = 11.09IPNFSDD258 pKa = 2.95NGEE261 pKa = 4.19FEE263 pKa = 4.41CVEE266 pKa = 4.08EE267 pKa = 5.34KK268 pKa = 10.82ILVDD272 pKa = 3.15KK273 pKa = 9.67TNPFGFFF280 pKa = 3.32
Molecular weight: 31.53 kDa
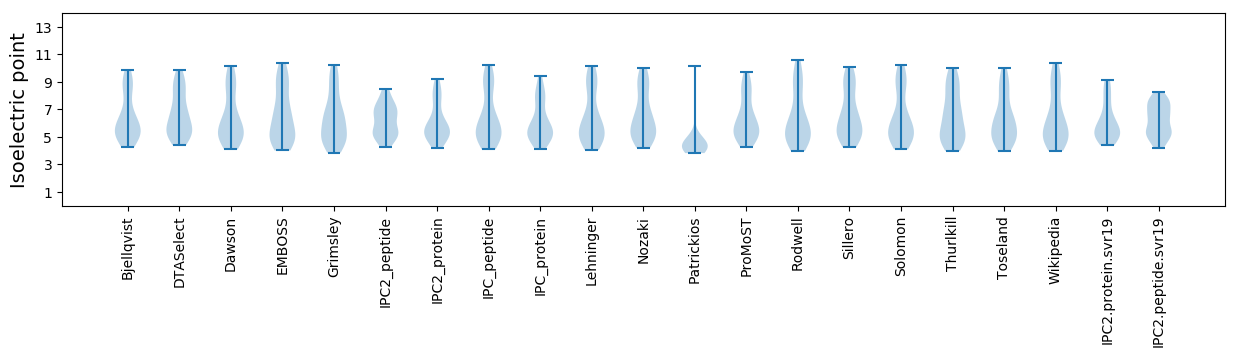
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223W0H2|A0A223W0H2_9CAUD Single stranded DNA-binding protein OS=Agrobacterium phage Atu_ph04 OX=2024263 PE=4 SV=1
MM1 pKa = 8.16AEE3 pKa = 4.07EE4 pKa = 4.9DD5 pKa = 3.84KK6 pKa = 11.23DD7 pKa = 3.88VQKK10 pKa = 9.93TFDD13 pKa = 4.43EE14 pKa = 4.45IVKK17 pKa = 10.21SLEE20 pKa = 3.91RR21 pKa = 11.84GMSSSEE27 pKa = 3.37IAKK30 pKa = 9.81RR31 pKa = 11.84QRR33 pKa = 11.84EE34 pKa = 4.34SIAYY38 pKa = 9.34FSGRR42 pKa = 11.84NNAKK46 pKa = 8.79TVDD49 pKa = 3.33TGRR52 pKa = 11.84ANLSMMTKK60 pKa = 9.48GQRR63 pKa = 11.84GVSDD67 pKa = 5.52LIPGTMMTYY76 pKa = 10.38AYY78 pKa = 9.73SAKK81 pKa = 10.27NAEE84 pKa = 4.32TLPYY88 pKa = 9.1WDD90 pKa = 4.86RR91 pKa = 11.84FPLIVFLDD99 pKa = 3.74VQKK102 pKa = 11.26NGNLLALNMHH112 pKa = 6.67YY113 pKa = 10.52LPPDD117 pKa = 3.82FRR119 pKa = 11.84AKK121 pKa = 10.14ILAEE125 pKa = 3.91LMKK128 pKa = 10.4TVTAKK133 pKa = 10.38EE134 pKa = 3.91LRR136 pKa = 11.84HH137 pKa = 5.69DD138 pKa = 3.22VRR140 pKa = 11.84MRR142 pKa = 11.84ITYY145 pKa = 7.48EE146 pKa = 3.43KK147 pKa = 9.14CQRR150 pKa = 11.84IAAFQPLQFALKK162 pKa = 10.1SYY164 pKa = 9.57IPQRR168 pKa = 11.84ISTKK172 pKa = 9.53IVRR175 pKa = 11.84IQPDD179 pKa = 3.27AWHH182 pKa = 6.55HH183 pKa = 6.5AIFFPSQMFVNASEE197 pKa = 4.24RR198 pKa = 11.84KK199 pKa = 7.8VWSDD203 pKa = 3.36FRR205 pKa = 11.84RR206 pKa = 11.84FRR208 pKa = 4.68
MM1 pKa = 8.16AEE3 pKa = 4.07EE4 pKa = 4.9DD5 pKa = 3.84KK6 pKa = 11.23DD7 pKa = 3.88VQKK10 pKa = 9.93TFDD13 pKa = 4.43EE14 pKa = 4.45IVKK17 pKa = 10.21SLEE20 pKa = 3.91RR21 pKa = 11.84GMSSSEE27 pKa = 3.37IAKK30 pKa = 9.81RR31 pKa = 11.84QRR33 pKa = 11.84EE34 pKa = 4.34SIAYY38 pKa = 9.34FSGRR42 pKa = 11.84NNAKK46 pKa = 8.79TVDD49 pKa = 3.33TGRR52 pKa = 11.84ANLSMMTKK60 pKa = 9.48GQRR63 pKa = 11.84GVSDD67 pKa = 5.52LIPGTMMTYY76 pKa = 10.38AYY78 pKa = 9.73SAKK81 pKa = 10.27NAEE84 pKa = 4.32TLPYY88 pKa = 9.1WDD90 pKa = 4.86RR91 pKa = 11.84FPLIVFLDD99 pKa = 3.74VQKK102 pKa = 11.26NGNLLALNMHH112 pKa = 6.67YY113 pKa = 10.52LPPDD117 pKa = 3.82FRR119 pKa = 11.84AKK121 pKa = 10.14ILAEE125 pKa = 3.91LMKK128 pKa = 10.4TVTAKK133 pKa = 10.38EE134 pKa = 3.91LRR136 pKa = 11.84HH137 pKa = 5.69DD138 pKa = 3.22VRR140 pKa = 11.84MRR142 pKa = 11.84ITYY145 pKa = 7.48EE146 pKa = 3.43KK147 pKa = 9.14CQRR150 pKa = 11.84IAAFQPLQFALKK162 pKa = 10.1SYY164 pKa = 9.57IPQRR168 pKa = 11.84ISTKK172 pKa = 9.53IVRR175 pKa = 11.84IQPDD179 pKa = 3.27AWHH182 pKa = 6.55HH183 pKa = 6.5AIFFPSQMFVNASEE197 pKa = 4.24RR198 pKa = 11.84KK199 pKa = 7.8VWSDD203 pKa = 3.36FRR205 pKa = 11.84RR206 pKa = 11.84FRR208 pKa = 4.68
Molecular weight: 24.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
19150 |
105 |
1695 |
399.0 |
44.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.313 ± 0.253 | 0.768 ± 0.078 |
6.695 ± 0.166 | 7.368 ± 0.423 |
5.097 ± 0.201 | 6.559 ± 0.459 |
1.749 ± 0.173 | 6.71 ± 0.23 |
6.971 ± 0.359 | 7.54 ± 0.267 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.136 ± 0.128 | 5.441 ± 0.209 |
3.603 ± 0.179 | 2.94 ± 0.124 |
5.196 ± 0.188 | 7.661 ± 0.379 |
5.676 ± 0.342 | 7.097 ± 0.29 |
1.05 ± 0.083 | 3.431 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
