
Halobacillus halophilus (strain ATCC 35676 / DSM 2266 / JCM 20832 / KCTC 3685 / LMG 17431 / NBRC 102448 / NCIMB 2269) (Sporosarcina halophila)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Halobacillus; Halobacillus halophilus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
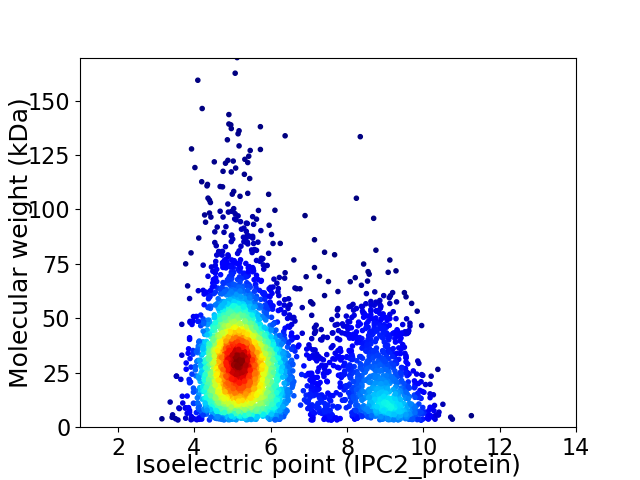
Virtual 2D-PAGE plot for 4100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0JI36|I0JI36_HALH3 Exopolysaccharide biosynthesis protein OS=Halobacillus halophilus (strain ATCC 35676 / DSM 2266 / JCM 20832 / KCTC 3685 / LMG 17431 / NBRC 102448 / NCIMB 2269) OX=866895 GN=HBHAL_1433 PE=3 SV=1
MM1 pKa = 7.21EE2 pKa = 6.15WIRR5 pKa = 11.84LNPDD9 pKa = 2.53ICSLFSNDD17 pKa = 3.89SVLVCYY23 pKa = 9.26LTDD26 pKa = 3.55EE27 pKa = 4.73LGEE30 pKa = 4.25TVEE33 pKa = 5.66SITCRR38 pKa = 11.84EE39 pKa = 3.99SGSRR43 pKa = 11.84EE44 pKa = 4.05DD45 pKa = 3.42VTVTLSGEE53 pKa = 4.38SVVLQRR59 pKa = 11.84VSLINQGFVVVEE71 pKa = 4.08ISDD74 pKa = 5.25DD75 pKa = 3.67EE76 pKa = 4.76TTCITDD82 pKa = 4.69PIPFCFVEE90 pKa = 4.28TLLLCAPDD98 pKa = 3.9GTDD101 pKa = 3.24IEE103 pKa = 4.89CKK105 pKa = 9.26VTEE108 pKa = 4.9FDD110 pKa = 4.24CQACVNCQNGEE121 pKa = 4.11FLSLDD126 pKa = 3.24IFFDD130 pKa = 3.63VCLSVQTVVDD140 pKa = 4.09TVFEE144 pKa = 4.36LPISFCVPRR153 pKa = 11.84KK154 pKa = 9.81EE155 pKa = 4.83IEE157 pKa = 4.07QPPCTFKK164 pKa = 10.84IPNQCPTTFSEE175 pKa = 5.36DD176 pKa = 3.19STEE179 pKa = 4.12RR180 pKa = 11.84KK181 pKa = 8.9QVQPEE186 pKa = 3.99VNEE189 pKa = 4.04ILNLPAPQPTPSQQQEE205 pKa = 4.57VACVSATKK213 pKa = 10.52VYY215 pKa = 10.87DD216 pKa = 3.31WVVSQNSFRR225 pKa = 11.84ITRR228 pKa = 11.84NQADD232 pKa = 3.37ITFQCFPCEE241 pKa = 3.54VDD243 pKa = 4.02LFVPADD249 pKa = 3.71IFCTNVISGRR259 pKa = 11.84VVCATIPIEE268 pKa = 4.22GAEE271 pKa = 4.33VTLSGSPDD279 pKa = 3.16LVLYY283 pKa = 10.8DD284 pKa = 4.24MNPVTTDD291 pKa = 2.82EE292 pKa = 4.24NGYY295 pKa = 10.38FDD297 pKa = 4.62VNVEE301 pKa = 3.99IPEE304 pKa = 4.27DD305 pKa = 3.94TEE307 pKa = 4.19ATNVTIMAEE316 pKa = 4.07AIVRR320 pKa = 11.84SVVASASTEE329 pKa = 4.25TVVEE333 pKa = 4.5CPEE336 pKa = 4.52DD337 pKa = 3.42PCGIDD342 pKa = 3.77LFAPATITCDD352 pKa = 3.18DD353 pKa = 3.92FMDD356 pKa = 4.32GRR358 pKa = 11.84VRR360 pKa = 11.84CNGRR364 pKa = 11.84LIEE367 pKa = 4.42GALVTFDD374 pKa = 4.01SNAPGIVTFEE384 pKa = 4.14PPQIEE389 pKa = 4.29TDD391 pKa = 3.52SLGDD395 pKa = 3.37YY396 pKa = 7.51FTGVRR401 pKa = 11.84ITNGTAPQTVTLIASTTVEE420 pKa = 3.99GQMVSDD426 pKa = 3.98SVDD429 pKa = 3.25VTVNCPFDD437 pKa = 4.16ACILTINVPQLIDD450 pKa = 3.63CMAVITGNISCNGMGIEE467 pKa = 3.96GAEE470 pKa = 3.97IFFSDD475 pKa = 4.12FPEE478 pKa = 4.32DD479 pKa = 3.58VVTYY483 pKa = 9.12EE484 pKa = 4.31PNPAISDD491 pKa = 3.57INGDD495 pKa = 3.64FTTTIMVPEE504 pKa = 4.66GTSLTDD510 pKa = 3.14IDD512 pKa = 4.62VEE514 pKa = 4.3ATTTVQGQMVEE525 pKa = 4.56ANFGTQIICLPEE537 pKa = 3.8EE538 pKa = 4.74CPCKK542 pKa = 10.39FRR544 pKa = 11.84IRR546 pKa = 11.84PRR548 pKa = 11.84GSRR551 pKa = 11.84TRR553 pKa = 11.84ATVEE557 pKa = 3.64LTQNGMVTFLDD568 pKa = 5.02GIINLSAIQCFRR580 pKa = 11.84IVPGCNPDD588 pKa = 3.12VDD590 pKa = 4.23DD591 pKa = 4.69FSVVFRR597 pKa = 11.84TDD599 pKa = 2.98QEE601 pKa = 4.29VIRR604 pKa = 11.84FVQGRR609 pKa = 11.84RR610 pKa = 11.84LDD612 pKa = 3.87IEE614 pKa = 4.59CEE616 pKa = 4.23SNTFARR622 pKa = 11.84VHH624 pKa = 5.38GTATAEE630 pKa = 4.12GNVLNGLFDD639 pKa = 3.65VTIEE643 pKa = 4.2LSITAPDD650 pKa = 3.15IGVWTVSATDD660 pKa = 3.29NMGNTFFTSFVQRR673 pKa = 11.84VSRR676 pKa = 11.84STFIGDD682 pKa = 4.78CDD684 pKa = 4.03DD685 pKa = 4.41APNN688 pKa = 3.56
MM1 pKa = 7.21EE2 pKa = 6.15WIRR5 pKa = 11.84LNPDD9 pKa = 2.53ICSLFSNDD17 pKa = 3.89SVLVCYY23 pKa = 9.26LTDD26 pKa = 3.55EE27 pKa = 4.73LGEE30 pKa = 4.25TVEE33 pKa = 5.66SITCRR38 pKa = 11.84EE39 pKa = 3.99SGSRR43 pKa = 11.84EE44 pKa = 4.05DD45 pKa = 3.42VTVTLSGEE53 pKa = 4.38SVVLQRR59 pKa = 11.84VSLINQGFVVVEE71 pKa = 4.08ISDD74 pKa = 5.25DD75 pKa = 3.67EE76 pKa = 4.76TTCITDD82 pKa = 4.69PIPFCFVEE90 pKa = 4.28TLLLCAPDD98 pKa = 3.9GTDD101 pKa = 3.24IEE103 pKa = 4.89CKK105 pKa = 9.26VTEE108 pKa = 4.9FDD110 pKa = 4.24CQACVNCQNGEE121 pKa = 4.11FLSLDD126 pKa = 3.24IFFDD130 pKa = 3.63VCLSVQTVVDD140 pKa = 4.09TVFEE144 pKa = 4.36LPISFCVPRR153 pKa = 11.84KK154 pKa = 9.81EE155 pKa = 4.83IEE157 pKa = 4.07QPPCTFKK164 pKa = 10.84IPNQCPTTFSEE175 pKa = 5.36DD176 pKa = 3.19STEE179 pKa = 4.12RR180 pKa = 11.84KK181 pKa = 8.9QVQPEE186 pKa = 3.99VNEE189 pKa = 4.04ILNLPAPQPTPSQQQEE205 pKa = 4.57VACVSATKK213 pKa = 10.52VYY215 pKa = 10.87DD216 pKa = 3.31WVVSQNSFRR225 pKa = 11.84ITRR228 pKa = 11.84NQADD232 pKa = 3.37ITFQCFPCEE241 pKa = 3.54VDD243 pKa = 4.02LFVPADD249 pKa = 3.71IFCTNVISGRR259 pKa = 11.84VVCATIPIEE268 pKa = 4.22GAEE271 pKa = 4.33VTLSGSPDD279 pKa = 3.16LVLYY283 pKa = 10.8DD284 pKa = 4.24MNPVTTDD291 pKa = 2.82EE292 pKa = 4.24NGYY295 pKa = 10.38FDD297 pKa = 4.62VNVEE301 pKa = 3.99IPEE304 pKa = 4.27DD305 pKa = 3.94TEE307 pKa = 4.19ATNVTIMAEE316 pKa = 4.07AIVRR320 pKa = 11.84SVVASASTEE329 pKa = 4.25TVVEE333 pKa = 4.5CPEE336 pKa = 4.52DD337 pKa = 3.42PCGIDD342 pKa = 3.77LFAPATITCDD352 pKa = 3.18DD353 pKa = 3.92FMDD356 pKa = 4.32GRR358 pKa = 11.84VRR360 pKa = 11.84CNGRR364 pKa = 11.84LIEE367 pKa = 4.42GALVTFDD374 pKa = 4.01SNAPGIVTFEE384 pKa = 4.14PPQIEE389 pKa = 4.29TDD391 pKa = 3.52SLGDD395 pKa = 3.37YY396 pKa = 7.51FTGVRR401 pKa = 11.84ITNGTAPQTVTLIASTTVEE420 pKa = 3.99GQMVSDD426 pKa = 3.98SVDD429 pKa = 3.25VTVNCPFDD437 pKa = 4.16ACILTINVPQLIDD450 pKa = 3.63CMAVITGNISCNGMGIEE467 pKa = 3.96GAEE470 pKa = 3.97IFFSDD475 pKa = 4.12FPEE478 pKa = 4.32DD479 pKa = 3.58VVTYY483 pKa = 9.12EE484 pKa = 4.31PNPAISDD491 pKa = 3.57INGDD495 pKa = 3.64FTTTIMVPEE504 pKa = 4.66GTSLTDD510 pKa = 3.14IDD512 pKa = 4.62VEE514 pKa = 4.3ATTTVQGQMVEE525 pKa = 4.56ANFGTQIICLPEE537 pKa = 3.8EE538 pKa = 4.74CPCKK542 pKa = 10.39FRR544 pKa = 11.84IRR546 pKa = 11.84PRR548 pKa = 11.84GSRR551 pKa = 11.84TRR553 pKa = 11.84ATVEE557 pKa = 3.64LTQNGMVTFLDD568 pKa = 5.02GIINLSAIQCFRR580 pKa = 11.84IVPGCNPDD588 pKa = 3.12VDD590 pKa = 4.23DD591 pKa = 4.69FSVVFRR597 pKa = 11.84TDD599 pKa = 2.98QEE601 pKa = 4.29VIRR604 pKa = 11.84FVQGRR609 pKa = 11.84RR610 pKa = 11.84LDD612 pKa = 3.87IEE614 pKa = 4.59CEE616 pKa = 4.23SNTFARR622 pKa = 11.84VHH624 pKa = 5.38GTATAEE630 pKa = 4.12GNVLNGLFDD639 pKa = 3.65VTIEE643 pKa = 4.2LSITAPDD650 pKa = 3.15IGVWTVSATDD660 pKa = 3.29NMGNTFFTSFVQRR673 pKa = 11.84VSRR676 pKa = 11.84STFIGDD682 pKa = 4.78CDD684 pKa = 4.03DD685 pKa = 4.41APNN688 pKa = 3.56
Molecular weight: 75.02 kDa
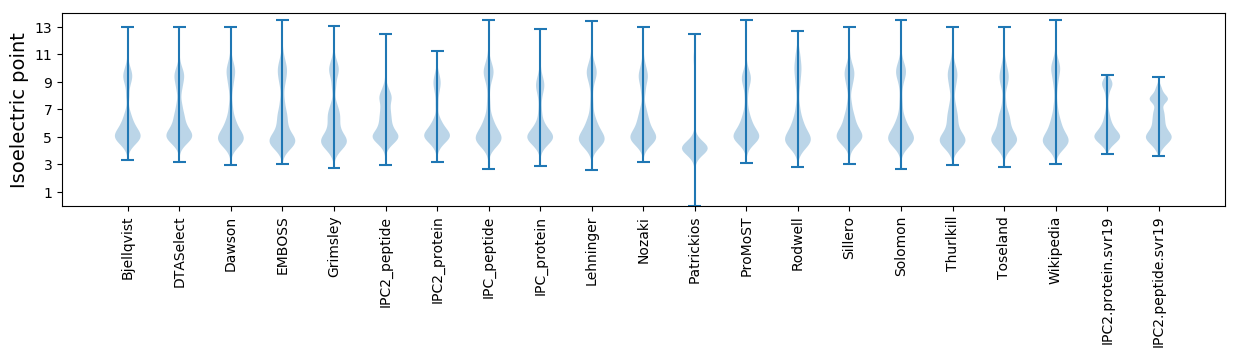
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0JH13|I0JH13_HALH3 DNA polymerase III delta' subunit OS=Halobacillus halophilus (strain ATCC 35676 / DSM 2266 / JCM 20832 / KCTC 3685 / LMG 17431 / NBRC 102448 / NCIMB 2269) OX=866895 GN=holB PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSSPNGRR28 pKa = 11.84KK29 pKa = 9.21VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSSPNGRR28 pKa = 11.84KK29 pKa = 9.21VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128406 |
30 |
1526 |
275.2 |
30.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.866 ± 0.042 | 0.623 ± 0.011 |
5.307 ± 0.036 | 8.053 ± 0.054 |
4.494 ± 0.034 | 6.981 ± 0.046 |
2.285 ± 0.021 | 7.153 ± 0.04 |
6.329 ± 0.039 | 9.56 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.919 ± 0.018 | 4.182 ± 0.027 |
3.745 ± 0.021 | 3.995 ± 0.028 |
4.244 ± 0.031 | 6.309 ± 0.03 |
5.318 ± 0.028 | 6.95 ± 0.033 |
1.1 ± 0.017 | 3.588 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
