
Halorubrum vacuolatum (Natronobacterium vacuolatum)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum
Average proteome isoelectric point is 5.0
Get precalculated fractions of proteins
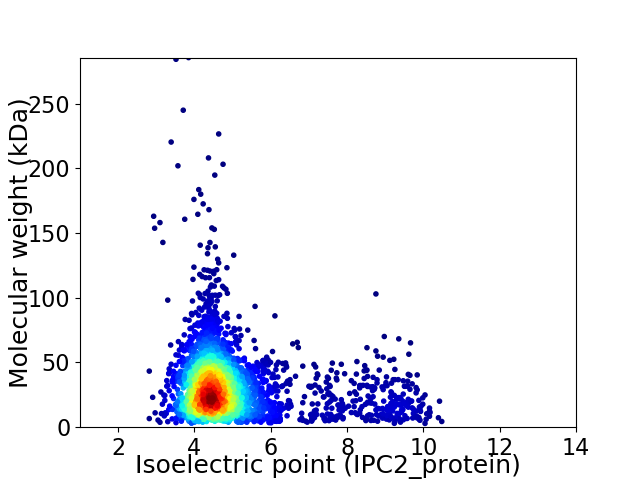
Virtual 2D-PAGE plot for 3371 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
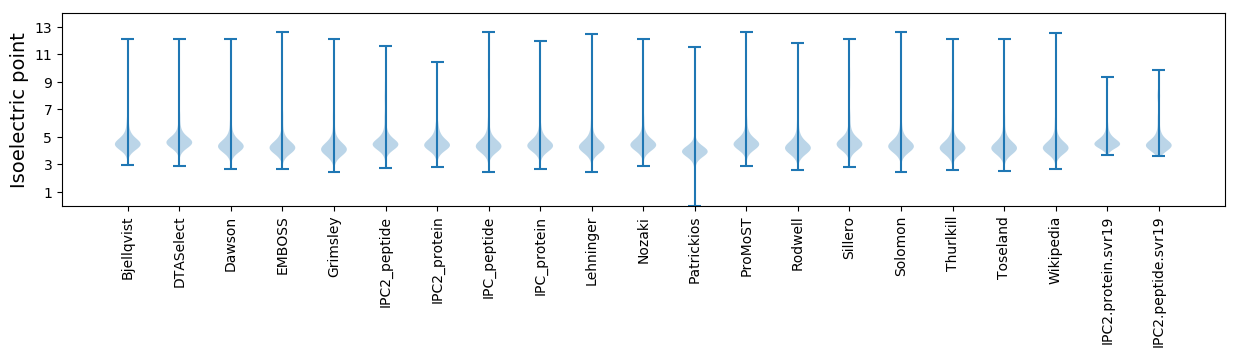
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238W4I5|A0A238W4I5_HALVU Dimethylglycine oxidase OS=Halorubrum vacuolatum OX=63740 GN=SAMN06264855_105105 PE=3 SV=1
MM1 pKa = 7.54FPRR4 pKa = 11.84EE5 pKa = 3.92YY6 pKa = 10.29RR7 pKa = 11.84GVAFAVALFVLVQLGALALVPSFAEE32 pKa = 3.89SGYY35 pKa = 10.73QPVDD39 pKa = 3.71DD40 pKa = 5.12PQDD43 pKa = 3.72PVNSLIYY50 pKa = 9.85IVAIVVMTGFMLAAFRR66 pKa = 11.84YY67 pKa = 8.38EE68 pKa = 4.12LDD70 pKa = 2.84WAIRR74 pKa = 11.84LLIVGVSGWLSWYY87 pKa = 9.65VFSAILPPLLAAIPAIAVAVGLLVHH112 pKa = 6.31PEE114 pKa = 3.7WYY116 pKa = 10.47VIDD119 pKa = 3.83SAGVLMGAGAAGLFGITFGILPALILLVFLAIYY152 pKa = 9.95DD153 pKa = 4.47AISVYY158 pKa = 9.3GTEE161 pKa = 4.41HH162 pKa = 6.88MLTLAEE168 pKa = 4.67GVMDD172 pKa = 4.42LNIPVVLVIPTTLSYY187 pKa = 11.5SLLEE191 pKa = 4.81DD192 pKa = 4.5DD193 pKa = 5.41FSGAADD199 pKa = 3.4VHH201 pKa = 6.56EE202 pKa = 5.21GDD204 pKa = 5.01DD205 pKa = 3.72EE206 pKa = 5.0GEE208 pKa = 3.87EE209 pKa = 4.41GEE211 pKa = 4.65PIKK214 pKa = 10.89GDD216 pKa = 3.32EE217 pKa = 4.73DD218 pKa = 4.64GEE220 pKa = 4.34ITADD224 pKa = 3.54SDD226 pKa = 4.9DD227 pKa = 5.3EE228 pKa = 4.24ITADD232 pKa = 5.0SDD234 pKa = 4.81DD235 pKa = 3.98EE236 pKa = 5.96HH237 pKa = 7.03DD238 pKa = 4.23TADD241 pKa = 4.74GDD243 pKa = 5.7DD244 pKa = 4.11PDD246 pKa = 4.73EE247 pKa = 5.07DD248 pKa = 3.95PSRR251 pKa = 11.84DD252 pKa = 3.29AFFIGLGDD260 pKa = 3.74AVIPAVLVASAATFSPAPHH279 pKa = 6.7LSVPVVGLNLPALLAMVGIVAGLLVLMSWVMKK311 pKa = 10.23GRR313 pKa = 11.84AHH315 pKa = 7.57AGLPLLNGGAIGGYY329 pKa = 10.39LIGSLLAGVPLIEE342 pKa = 4.71AVGLGPFVV350 pKa = 3.38
MM1 pKa = 7.54FPRR4 pKa = 11.84EE5 pKa = 3.92YY6 pKa = 10.29RR7 pKa = 11.84GVAFAVALFVLVQLGALALVPSFAEE32 pKa = 3.89SGYY35 pKa = 10.73QPVDD39 pKa = 3.71DD40 pKa = 5.12PQDD43 pKa = 3.72PVNSLIYY50 pKa = 9.85IVAIVVMTGFMLAAFRR66 pKa = 11.84YY67 pKa = 8.38EE68 pKa = 4.12LDD70 pKa = 2.84WAIRR74 pKa = 11.84LLIVGVSGWLSWYY87 pKa = 9.65VFSAILPPLLAAIPAIAVAVGLLVHH112 pKa = 6.31PEE114 pKa = 3.7WYY116 pKa = 10.47VIDD119 pKa = 3.83SAGVLMGAGAAGLFGITFGILPALILLVFLAIYY152 pKa = 9.95DD153 pKa = 4.47AISVYY158 pKa = 9.3GTEE161 pKa = 4.41HH162 pKa = 6.88MLTLAEE168 pKa = 4.67GVMDD172 pKa = 4.42LNIPVVLVIPTTLSYY187 pKa = 11.5SLLEE191 pKa = 4.81DD192 pKa = 4.5DD193 pKa = 5.41FSGAADD199 pKa = 3.4VHH201 pKa = 6.56EE202 pKa = 5.21GDD204 pKa = 5.01DD205 pKa = 3.72EE206 pKa = 5.0GEE208 pKa = 3.87EE209 pKa = 4.41GEE211 pKa = 4.65PIKK214 pKa = 10.89GDD216 pKa = 3.32EE217 pKa = 4.73DD218 pKa = 4.64GEE220 pKa = 4.34ITADD224 pKa = 3.54SDD226 pKa = 4.9DD227 pKa = 5.3EE228 pKa = 4.24ITADD232 pKa = 5.0SDD234 pKa = 4.81DD235 pKa = 3.98EE236 pKa = 5.96HH237 pKa = 7.03DD238 pKa = 4.23TADD241 pKa = 4.74GDD243 pKa = 5.7DD244 pKa = 4.11PDD246 pKa = 4.73EE247 pKa = 5.07DD248 pKa = 3.95PSRR251 pKa = 11.84DD252 pKa = 3.29AFFIGLGDD260 pKa = 3.74AVIPAVLVASAATFSPAPHH279 pKa = 6.7LSVPVVGLNLPALLAMVGIVAGLLVLMSWVMKK311 pKa = 10.23GRR313 pKa = 11.84AHH315 pKa = 7.57AGLPLLNGGAIGGYY329 pKa = 10.39LIGSLLAGVPLIEE342 pKa = 4.71AVGLGPFVV350 pKa = 3.38
Molecular weight: 36.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A238VL29|A0A238VL29_HALVU C2H2-type zinc finger OS=Halorubrum vacuolatum OX=63740 GN=SAMN06264855_10358 PE=4 SV=1
MM1 pKa = 7.36ATLLCRR7 pKa = 11.84RR8 pKa = 11.84CFPGFFSEE16 pKa = 5.14LPTGSGDD23 pKa = 3.23RR24 pKa = 11.84SIRR27 pKa = 11.84YY28 pKa = 8.26NFSQCLVLEE37 pKa = 4.32YY38 pKa = 10.95SCIRR42 pKa = 11.84RR43 pKa = 11.84FALHH47 pKa = 6.69LVKK50 pKa = 10.6LPNVRR55 pKa = 11.84APSSVSQHH63 pKa = 5.84TIYY66 pKa = 10.84SIGPGQNLLDD76 pKa = 4.64GIKK79 pKa = 10.28QIVRR83 pKa = 11.84GHH85 pKa = 5.29TLSTEE90 pKa = 3.94NTGGNRR96 pKa = 11.84LARR99 pKa = 11.84DD100 pKa = 3.68GVSAPKK106 pKa = 10.42LDD108 pKa = 3.78VKK110 pKa = 8.69YY111 pKa = 10.34HH112 pKa = 6.25RR113 pKa = 11.84SHH115 pKa = 7.19RR116 pKa = 11.84PTASPPKK123 pKa = 9.33SHH125 pKa = 5.5IHH127 pKa = 5.07GRR129 pKa = 11.84AVATSIKK136 pKa = 9.46CTIPNAPHH144 pKa = 6.99RR145 pKa = 11.84FSNGSDD151 pKa = 3.29GYY153 pKa = 9.72TIDD156 pKa = 5.59LRR158 pKa = 11.84QQAAGNTARR167 pKa = 11.84EE168 pKa = 4.03SDD170 pKa = 3.46KK171 pKa = 11.37LGADD175 pKa = 3.05RR176 pKa = 11.84RR177 pKa = 11.84VITGG181 pKa = 3.32
MM1 pKa = 7.36ATLLCRR7 pKa = 11.84RR8 pKa = 11.84CFPGFFSEE16 pKa = 5.14LPTGSGDD23 pKa = 3.23RR24 pKa = 11.84SIRR27 pKa = 11.84YY28 pKa = 8.26NFSQCLVLEE37 pKa = 4.32YY38 pKa = 10.95SCIRR42 pKa = 11.84RR43 pKa = 11.84FALHH47 pKa = 6.69LVKK50 pKa = 10.6LPNVRR55 pKa = 11.84APSSVSQHH63 pKa = 5.84TIYY66 pKa = 10.84SIGPGQNLLDD76 pKa = 4.64GIKK79 pKa = 10.28QIVRR83 pKa = 11.84GHH85 pKa = 5.29TLSTEE90 pKa = 3.94NTGGNRR96 pKa = 11.84LARR99 pKa = 11.84DD100 pKa = 3.68GVSAPKK106 pKa = 10.42LDD108 pKa = 3.78VKK110 pKa = 8.69YY111 pKa = 10.34HH112 pKa = 6.25RR113 pKa = 11.84SHH115 pKa = 7.19RR116 pKa = 11.84PTASPPKK123 pKa = 9.33SHH125 pKa = 5.5IHH127 pKa = 5.07GRR129 pKa = 11.84AVATSIKK136 pKa = 9.46CTIPNAPHH144 pKa = 6.99RR145 pKa = 11.84FSNGSDD151 pKa = 3.29GYY153 pKa = 9.72TIDD156 pKa = 5.59LRR158 pKa = 11.84QQAAGNTARR167 pKa = 11.84EE168 pKa = 4.03SDD170 pKa = 3.46KK171 pKa = 11.37LGADD175 pKa = 3.05RR176 pKa = 11.84RR177 pKa = 11.84VITGG181 pKa = 3.32
Molecular weight: 19.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1002170 |
26 |
2727 |
297.3 |
32.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.899 ± 0.067 | 0.716 ± 0.014 |
8.415 ± 0.06 | 8.695 ± 0.061 |
3.298 ± 0.03 | 8.474 ± 0.046 |
2.062 ± 0.023 | 4.664 ± 0.032 |
1.947 ± 0.034 | 8.727 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.71 ± 0.021 | 2.307 ± 0.027 |
4.746 ± 0.037 | 2.217 ± 0.029 |
6.924 ± 0.046 | 5.241 ± 0.032 |
6.499 ± 0.047 | 8.723 ± 0.04 |
1.103 ± 0.018 | 2.633 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
