
Mesorhizobium carbonis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
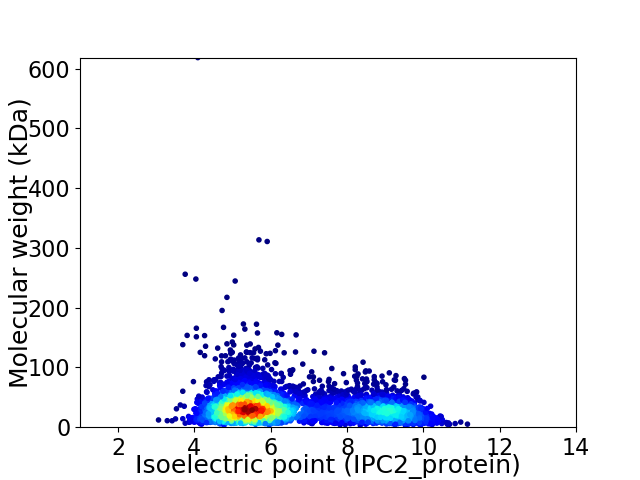
Virtual 2D-PAGE plot for 4987 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
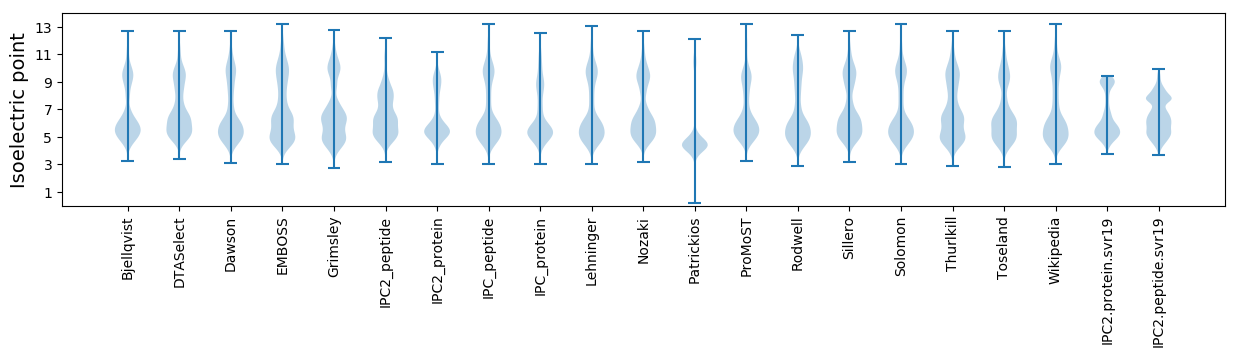
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A429Z023|A0A429Z023_9RHIZ Lipopolysaccharide biosynthesis protein OS=Mesorhizobium carbonis OX=2495581 GN=EJC49_07325 PE=4 SV=1
MM1 pKa = 7.58DD2 pKa = 3.33QRR4 pKa = 11.84RR5 pKa = 11.84TTRR8 pKa = 11.84ALLSGAALGTVFLALGMPAPAEE30 pKa = 4.1AACLADD36 pKa = 4.44GTTLTCTGDD45 pKa = 3.33LGAGVIVADD54 pKa = 4.4PIEE57 pKa = 4.17RR58 pKa = 11.84LEE60 pKa = 4.25INGVTAMIAPAEE72 pKa = 4.24GVSGIDD78 pKa = 3.69FQSSGVIAIVSDD90 pKa = 3.52TGAYY94 pKa = 9.62QILATDD100 pKa = 3.82ADD102 pKa = 4.99GIHH105 pKa = 6.78ADD107 pKa = 3.4SWNGSITIEE116 pKa = 3.95HH117 pKa = 6.45TGDD120 pKa = 2.9IRR122 pKa = 11.84SVGGYY127 pKa = 7.58GVRR130 pKa = 11.84TISLAGTSVTVDD142 pKa = 3.13GDD144 pKa = 3.71IEE146 pKa = 4.62ASLGGIVVNNDD157 pKa = 3.26DD158 pKa = 4.67AGTTTEE164 pKa = 4.22ISHH167 pKa = 6.51TGDD170 pKa = 2.93ILVTEE175 pKa = 4.79GAGISAVTSSTPLVVVNEE193 pKa = 4.05GSINASGDD201 pKa = 3.38GISAVNTSEE210 pKa = 4.23SGSVTVTQTGNVIAGDD226 pKa = 3.82RR227 pKa = 11.84GIHH230 pKa = 6.42ADD232 pKa = 3.34AAFGSVTLTSAGNISSVGDD251 pKa = 4.34GIQLLSTGSDD261 pKa = 3.06VTATHH266 pKa = 6.94TGNVTSTEE274 pKa = 3.91GRR276 pKa = 11.84GIFLDD281 pKa = 3.97APTAGASLSGSGTIQSYY298 pKa = 11.04GDD300 pKa = 3.76GIFAQSTGVAHH311 pKa = 5.9STSVTWTGNITSEE324 pKa = 3.93IGKK327 pKa = 10.21GIYY330 pKa = 10.01AYY332 pKa = 10.05AANGAVTVLSAGDD345 pKa = 3.61IVSSGDD351 pKa = 3.42SIFAMNVSTDD361 pKa = 3.11GTAEE365 pKa = 4.13DD366 pKa = 4.07VSVTHH371 pKa = 6.7TGDD374 pKa = 3.21LTSNAGNGVYY384 pKa = 10.14AYY386 pKa = 8.17TAGGQVDD393 pKa = 3.84VTVNNSILTASGYY406 pKa = 11.16GIFAQTKK413 pKa = 7.66GTAVDD418 pKa = 4.01DD419 pKa = 4.39DD420 pKa = 4.17VTVEE424 pKa = 4.03FNGTIAQSWIGILAEE439 pKa = 4.49ANTGTVLVTSSGEE452 pKa = 3.58IAAWDD457 pKa = 3.66HH458 pKa = 6.55GIWAANTGGGNVTVSQTGGIDD479 pKa = 3.0AGTGHH484 pKa = 8.04AIYY487 pKa = 9.83AYY489 pKa = 9.07SPHH492 pKa = 6.45GAVSVTATGDD502 pKa = 3.35MFRR505 pKa = 11.84SGMTAFYY512 pKa = 11.21VEE514 pKa = 4.61TKK516 pKa = 10.06SGNAATVNVTGDD528 pKa = 3.25IVSGDD533 pKa = 3.53GGITALSAQGTVTVTMRR550 pKa = 11.84GDD552 pKa = 2.99IDD554 pKa = 3.89AGADD558 pKa = 3.75GIYY561 pKa = 10.7AEE563 pKa = 5.23SKK565 pKa = 11.1DD566 pKa = 3.65NDD568 pKa = 3.92LVKK571 pKa = 10.64VDD573 pKa = 4.31TIGDD577 pKa = 3.54ITAGGSAIFAASSQGVVDD595 pKa = 3.87VDD597 pKa = 4.6SIGNLSAGGYY607 pKa = 9.88GIYY610 pKa = 10.26AANLGDD616 pKa = 3.85NVVTVNSTGNIDD628 pKa = 3.21AGNRR632 pKa = 11.84GIHH635 pKa = 5.86AQSATQAVTIVTSGAVTSGSDD656 pKa = 3.16AVFAEE661 pKa = 4.3SLGGNIVSITSIGNLVAGTDD681 pKa = 3.38GDD683 pKa = 4.2GIYY686 pKa = 10.7AFSATGTVTVTSTGDD701 pKa = 2.8IDD703 pKa = 5.32AGDD706 pKa = 4.26DD707 pKa = 4.22GIFARR712 pKa = 11.84STGGQDD718 pKa = 3.44VSVTSLGDD726 pKa = 3.05IGAGTGYY733 pKa = 10.87GIYY736 pKa = 10.27GYY738 pKa = 10.05SATGAVTIGSTGEE751 pKa = 4.14IVSGDD756 pKa = 3.34HH757 pKa = 7.53GIYY760 pKa = 10.52ALNLGYY766 pKa = 8.96GTVGVTQVGDD776 pKa = 3.29IDD778 pKa = 4.36AGGSGIRR785 pKa = 11.84ATSTAGIVSVSNKK798 pKa = 10.01GNIDD802 pKa = 3.38AGGDD806 pKa = 3.6GIYY809 pKa = 10.83ALTLGYY815 pKa = 9.94EE816 pKa = 4.21AATVTNEE823 pKa = 4.24GYY825 pKa = 10.19IDD827 pKa = 3.71ANGYY831 pKa = 9.23GIHH834 pKa = 6.01GHH836 pKa = 6.05SDD838 pKa = 3.11AGSVTIDD845 pKa = 3.17NKK847 pKa = 9.74GAIWSGSDD855 pKa = 3.75GIRR858 pKa = 11.84ATTLSSSLVKK868 pKa = 9.58VTQLGDD874 pKa = 3.45IDD876 pKa = 3.92SGGYY880 pKa = 9.89GVFVNAEE887 pKa = 4.17AGDD890 pKa = 3.96ADD892 pKa = 4.17IDD894 pKa = 3.99TTGDD898 pKa = 3.12ITSVNHH904 pKa = 7.0GIYY907 pKa = 10.73VLTNGNTLIDD917 pKa = 3.59IDD919 pKa = 4.02HH920 pKa = 7.08RR921 pKa = 11.84GSITSTSGNGITATSVAQATIDD943 pKa = 3.59VSVSGGTVSGALDD956 pKa = 4.83GIRR959 pKa = 11.84LASEE963 pKa = 4.21RR964 pKa = 11.84EE965 pKa = 3.83MTVTVGADD973 pKa = 3.05ASVIGGAGNAGVRR986 pKa = 11.84FVEE989 pKa = 4.3GLGIQLTNRR998 pKa = 11.84GSISNAGGIDD1008 pKa = 2.92EE1009 pKa = 4.26WAIVSAQNDD1018 pKa = 4.02TTVDD1022 pKa = 3.26NYY1024 pKa = 10.19GTIAGNVLLGPWSNPFNNFAGALFNMGSTVNIGADD1059 pKa = 3.37STLSNHH1065 pKa = 5.77GTLSPGGEE1073 pKa = 3.84GVVQTTTLTGILVNEE1088 pKa = 4.26ATGTLLFDD1096 pKa = 3.84VDD1098 pKa = 4.08IEE1100 pKa = 4.27NATTDD1105 pKa = 3.63RR1106 pKa = 11.84LNVTNTAALDD1116 pKa = 3.65GNLRR1120 pKa = 11.84LNFVSATGTPGSYY1133 pKa = 10.2TIVTTGEE1140 pKa = 4.4GVTSQSLTLLNPFVLADD1157 pKa = 3.15IATVNNGNDD1166 pKa = 3.36VEE1168 pKa = 4.37LSIHH1172 pKa = 6.33GFDD1175 pKa = 4.85FSPAGMSEE1183 pKa = 4.05NASSIGNAIQTSIQGNGALEE1203 pKa = 4.72PIAVALLNLQTPEE1216 pKa = 3.84EE1217 pKa = 4.53AEE1219 pKa = 4.14DD1220 pKa = 3.87AFEE1223 pKa = 3.92QLSPNTYY1230 pKa = 9.77VADD1233 pKa = 3.8QIGSVDD1239 pKa = 4.33SIATFSNSMLSCRR1252 pKa = 11.84MASGEE1257 pKa = 4.09NAFGAEE1263 pKa = 4.92GEE1265 pKa = 4.71CAWGRR1270 pKa = 11.84AVYY1273 pKa = 9.94SAYY1276 pKa = 10.74DD1277 pKa = 3.55LAANGDD1283 pKa = 3.53NSGFNSRR1290 pKa = 11.84SVEE1293 pKa = 3.93MMGGVQVAVPDD1304 pKa = 4.03TAWRR1308 pKa = 11.84LGGSVGFQASEE1319 pKa = 3.62QDD1321 pKa = 3.7GNNGSSSEE1329 pKa = 4.07GSSFTVGAVAKK1340 pKa = 8.05YY1341 pKa = 10.74APGPYY1346 pKa = 10.05LFAASVAASRR1356 pKa = 11.84GNYY1359 pKa = 7.69DD1360 pKa = 3.39TLRR1363 pKa = 11.84PIDD1366 pKa = 3.76IGNFTDD1372 pKa = 4.07LLSGEE1377 pKa = 4.39TDD1379 pKa = 3.16VTTVSGRR1386 pKa = 11.84LRR1388 pKa = 11.84AAYY1391 pKa = 9.25TMEE1394 pKa = 3.61QGAFYY1399 pKa = 10.22LRR1401 pKa = 11.84PMVDD1405 pKa = 3.35ASATYY1410 pKa = 10.49VRR1412 pKa = 11.84TGAFTEE1418 pKa = 4.4TGGVASLSVGAVEE1431 pKa = 4.21NTVLALMPAIEE1442 pKa = 4.55IGGQVSVGEE1451 pKa = 4.3DD1452 pKa = 3.17MLLRR1456 pKa = 11.84PYY1458 pKa = 10.81LRR1460 pKa = 11.84GGASVYY1466 pKa = 10.76SGNEE1470 pKa = 3.56YY1471 pKa = 10.94SLTGSFTADD1480 pKa = 2.98GNAAAPFTVTSSSEE1494 pKa = 3.99DD1495 pKa = 3.6VLWTVSTGVDD1505 pKa = 3.77LLRR1508 pKa = 11.84GDD1510 pKa = 3.86VGTLQIFYY1518 pKa = 9.82EE1519 pKa = 4.88GAFGEE1524 pKa = 4.53HH1525 pKa = 5.02TTINSGGAKK1534 pKa = 10.27FSVNFF1539 pKa = 3.9
MM1 pKa = 7.58DD2 pKa = 3.33QRR4 pKa = 11.84RR5 pKa = 11.84TTRR8 pKa = 11.84ALLSGAALGTVFLALGMPAPAEE30 pKa = 4.1AACLADD36 pKa = 4.44GTTLTCTGDD45 pKa = 3.33LGAGVIVADD54 pKa = 4.4PIEE57 pKa = 4.17RR58 pKa = 11.84LEE60 pKa = 4.25INGVTAMIAPAEE72 pKa = 4.24GVSGIDD78 pKa = 3.69FQSSGVIAIVSDD90 pKa = 3.52TGAYY94 pKa = 9.62QILATDD100 pKa = 3.82ADD102 pKa = 4.99GIHH105 pKa = 6.78ADD107 pKa = 3.4SWNGSITIEE116 pKa = 3.95HH117 pKa = 6.45TGDD120 pKa = 2.9IRR122 pKa = 11.84SVGGYY127 pKa = 7.58GVRR130 pKa = 11.84TISLAGTSVTVDD142 pKa = 3.13GDD144 pKa = 3.71IEE146 pKa = 4.62ASLGGIVVNNDD157 pKa = 3.26DD158 pKa = 4.67AGTTTEE164 pKa = 4.22ISHH167 pKa = 6.51TGDD170 pKa = 2.93ILVTEE175 pKa = 4.79GAGISAVTSSTPLVVVNEE193 pKa = 4.05GSINASGDD201 pKa = 3.38GISAVNTSEE210 pKa = 4.23SGSVTVTQTGNVIAGDD226 pKa = 3.82RR227 pKa = 11.84GIHH230 pKa = 6.42ADD232 pKa = 3.34AAFGSVTLTSAGNISSVGDD251 pKa = 4.34GIQLLSTGSDD261 pKa = 3.06VTATHH266 pKa = 6.94TGNVTSTEE274 pKa = 3.91GRR276 pKa = 11.84GIFLDD281 pKa = 3.97APTAGASLSGSGTIQSYY298 pKa = 11.04GDD300 pKa = 3.76GIFAQSTGVAHH311 pKa = 5.9STSVTWTGNITSEE324 pKa = 3.93IGKK327 pKa = 10.21GIYY330 pKa = 10.01AYY332 pKa = 10.05AANGAVTVLSAGDD345 pKa = 3.61IVSSGDD351 pKa = 3.42SIFAMNVSTDD361 pKa = 3.11GTAEE365 pKa = 4.13DD366 pKa = 4.07VSVTHH371 pKa = 6.7TGDD374 pKa = 3.21LTSNAGNGVYY384 pKa = 10.14AYY386 pKa = 8.17TAGGQVDD393 pKa = 3.84VTVNNSILTASGYY406 pKa = 11.16GIFAQTKK413 pKa = 7.66GTAVDD418 pKa = 4.01DD419 pKa = 4.39DD420 pKa = 4.17VTVEE424 pKa = 4.03FNGTIAQSWIGILAEE439 pKa = 4.49ANTGTVLVTSSGEE452 pKa = 3.58IAAWDD457 pKa = 3.66HH458 pKa = 6.55GIWAANTGGGNVTVSQTGGIDD479 pKa = 3.0AGTGHH484 pKa = 8.04AIYY487 pKa = 9.83AYY489 pKa = 9.07SPHH492 pKa = 6.45GAVSVTATGDD502 pKa = 3.35MFRR505 pKa = 11.84SGMTAFYY512 pKa = 11.21VEE514 pKa = 4.61TKK516 pKa = 10.06SGNAATVNVTGDD528 pKa = 3.25IVSGDD533 pKa = 3.53GGITALSAQGTVTVTMRR550 pKa = 11.84GDD552 pKa = 2.99IDD554 pKa = 3.89AGADD558 pKa = 3.75GIYY561 pKa = 10.7AEE563 pKa = 5.23SKK565 pKa = 11.1DD566 pKa = 3.65NDD568 pKa = 3.92LVKK571 pKa = 10.64VDD573 pKa = 4.31TIGDD577 pKa = 3.54ITAGGSAIFAASSQGVVDD595 pKa = 3.87VDD597 pKa = 4.6SIGNLSAGGYY607 pKa = 9.88GIYY610 pKa = 10.26AANLGDD616 pKa = 3.85NVVTVNSTGNIDD628 pKa = 3.21AGNRR632 pKa = 11.84GIHH635 pKa = 5.86AQSATQAVTIVTSGAVTSGSDD656 pKa = 3.16AVFAEE661 pKa = 4.3SLGGNIVSITSIGNLVAGTDD681 pKa = 3.38GDD683 pKa = 4.2GIYY686 pKa = 10.7AFSATGTVTVTSTGDD701 pKa = 2.8IDD703 pKa = 5.32AGDD706 pKa = 4.26DD707 pKa = 4.22GIFARR712 pKa = 11.84STGGQDD718 pKa = 3.44VSVTSLGDD726 pKa = 3.05IGAGTGYY733 pKa = 10.87GIYY736 pKa = 10.27GYY738 pKa = 10.05SATGAVTIGSTGEE751 pKa = 4.14IVSGDD756 pKa = 3.34HH757 pKa = 7.53GIYY760 pKa = 10.52ALNLGYY766 pKa = 8.96GTVGVTQVGDD776 pKa = 3.29IDD778 pKa = 4.36AGGSGIRR785 pKa = 11.84ATSTAGIVSVSNKK798 pKa = 10.01GNIDD802 pKa = 3.38AGGDD806 pKa = 3.6GIYY809 pKa = 10.83ALTLGYY815 pKa = 9.94EE816 pKa = 4.21AATVTNEE823 pKa = 4.24GYY825 pKa = 10.19IDD827 pKa = 3.71ANGYY831 pKa = 9.23GIHH834 pKa = 6.01GHH836 pKa = 6.05SDD838 pKa = 3.11AGSVTIDD845 pKa = 3.17NKK847 pKa = 9.74GAIWSGSDD855 pKa = 3.75GIRR858 pKa = 11.84ATTLSSSLVKK868 pKa = 9.58VTQLGDD874 pKa = 3.45IDD876 pKa = 3.92SGGYY880 pKa = 9.89GVFVNAEE887 pKa = 4.17AGDD890 pKa = 3.96ADD892 pKa = 4.17IDD894 pKa = 3.99TTGDD898 pKa = 3.12ITSVNHH904 pKa = 7.0GIYY907 pKa = 10.73VLTNGNTLIDD917 pKa = 3.59IDD919 pKa = 4.02HH920 pKa = 7.08RR921 pKa = 11.84GSITSTSGNGITATSVAQATIDD943 pKa = 3.59VSVSGGTVSGALDD956 pKa = 4.83GIRR959 pKa = 11.84LASEE963 pKa = 4.21RR964 pKa = 11.84EE965 pKa = 3.83MTVTVGADD973 pKa = 3.05ASVIGGAGNAGVRR986 pKa = 11.84FVEE989 pKa = 4.3GLGIQLTNRR998 pKa = 11.84GSISNAGGIDD1008 pKa = 2.92EE1009 pKa = 4.26WAIVSAQNDD1018 pKa = 4.02TTVDD1022 pKa = 3.26NYY1024 pKa = 10.19GTIAGNVLLGPWSNPFNNFAGALFNMGSTVNIGADD1059 pKa = 3.37STLSNHH1065 pKa = 5.77GTLSPGGEE1073 pKa = 3.84GVVQTTTLTGILVNEE1088 pKa = 4.26ATGTLLFDD1096 pKa = 3.84VDD1098 pKa = 4.08IEE1100 pKa = 4.27NATTDD1105 pKa = 3.63RR1106 pKa = 11.84LNVTNTAALDD1116 pKa = 3.65GNLRR1120 pKa = 11.84LNFVSATGTPGSYY1133 pKa = 10.2TIVTTGEE1140 pKa = 4.4GVTSQSLTLLNPFVLADD1157 pKa = 3.15IATVNNGNDD1166 pKa = 3.36VEE1168 pKa = 4.37LSIHH1172 pKa = 6.33GFDD1175 pKa = 4.85FSPAGMSEE1183 pKa = 4.05NASSIGNAIQTSIQGNGALEE1203 pKa = 4.72PIAVALLNLQTPEE1216 pKa = 3.84EE1217 pKa = 4.53AEE1219 pKa = 4.14DD1220 pKa = 3.87AFEE1223 pKa = 3.92QLSPNTYY1230 pKa = 9.77VADD1233 pKa = 3.8QIGSVDD1239 pKa = 4.33SIATFSNSMLSCRR1252 pKa = 11.84MASGEE1257 pKa = 4.09NAFGAEE1263 pKa = 4.92GEE1265 pKa = 4.71CAWGRR1270 pKa = 11.84AVYY1273 pKa = 9.94SAYY1276 pKa = 10.74DD1277 pKa = 3.55LAANGDD1283 pKa = 3.53NSGFNSRR1290 pKa = 11.84SVEE1293 pKa = 3.93MMGGVQVAVPDD1304 pKa = 4.03TAWRR1308 pKa = 11.84LGGSVGFQASEE1319 pKa = 3.62QDD1321 pKa = 3.7GNNGSSSEE1329 pKa = 4.07GSSFTVGAVAKK1340 pKa = 8.05YY1341 pKa = 10.74APGPYY1346 pKa = 10.05LFAASVAASRR1356 pKa = 11.84GNYY1359 pKa = 7.69DD1360 pKa = 3.39TLRR1363 pKa = 11.84PIDD1366 pKa = 3.76IGNFTDD1372 pKa = 4.07LLSGEE1377 pKa = 4.39TDD1379 pKa = 3.16VTTVSGRR1386 pKa = 11.84LRR1388 pKa = 11.84AAYY1391 pKa = 9.25TMEE1394 pKa = 3.61QGAFYY1399 pKa = 10.22LRR1401 pKa = 11.84PMVDD1405 pKa = 3.35ASATYY1410 pKa = 10.49VRR1412 pKa = 11.84TGAFTEE1418 pKa = 4.4TGGVASLSVGAVEE1431 pKa = 4.21NTVLALMPAIEE1442 pKa = 4.55IGGQVSVGEE1451 pKa = 4.3DD1452 pKa = 3.17MLLRR1456 pKa = 11.84PYY1458 pKa = 10.81LRR1460 pKa = 11.84GGASVYY1466 pKa = 10.76SGNEE1470 pKa = 3.56YY1471 pKa = 10.94SLTGSFTADD1480 pKa = 2.98GNAAAPFTVTSSSEE1494 pKa = 3.99DD1495 pKa = 3.6VLWTVSTGVDD1505 pKa = 3.77LLRR1508 pKa = 11.84GDD1510 pKa = 3.86VGTLQIFYY1518 pKa = 9.82EE1519 pKa = 4.88GAFGEE1524 pKa = 4.53HH1525 pKa = 5.02TTINSGGAKK1534 pKa = 10.27FSVNFF1539 pKa = 3.9
Molecular weight: 153.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A429YZ32|A0A429YZ32_9RHIZ Uncharacterized protein OS=Mesorhizobium carbonis OX=2495581 GN=EJC49_09135 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1586296 |
22 |
6283 |
318.1 |
34.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.77 ± 0.053 | 0.8 ± 0.012 |
5.88 ± 0.029 | 5.714 ± 0.036 |
3.844 ± 0.025 | 8.856 ± 0.049 |
2.037 ± 0.018 | 5.225 ± 0.027 |
2.883 ± 0.03 | 9.861 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.594 ± 0.018 | 2.402 ± 0.022 |
5.122 ± 0.029 | 2.801 ± 0.021 |
7.489 ± 0.044 | 5.358 ± 0.031 |
5.25 ± 0.03 | 7.648 ± 0.031 |
1.336 ± 0.015 | 2.131 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
