
Gammapapillomavirus 18
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
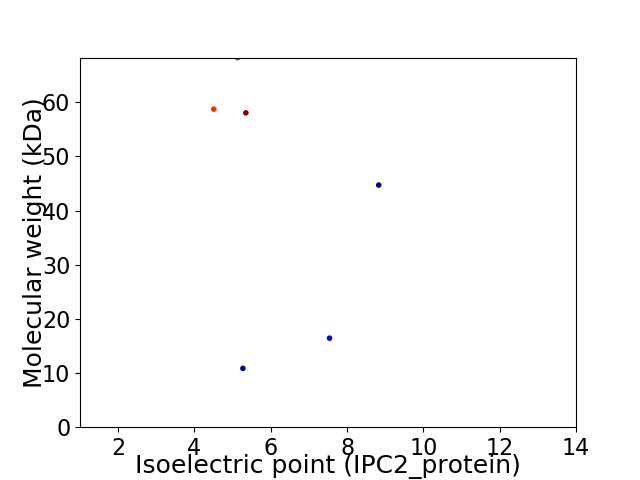
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2D2AM25|A0A2D2AM25_9PAPI Minor capsid protein L2 OS=Gammapapillomavirus 18 OX=1513263 GN=L2 PE=3 SV=1
MM1 pKa = 7.18SVSRR5 pKa = 11.84NKK7 pKa = 10.32RR8 pKa = 11.84DD9 pKa = 3.62TVEE12 pKa = 4.29NLYY15 pKa = 10.59KK16 pKa = 10.31SCKK19 pKa = 10.05LGGDD23 pKa = 3.95CPPDD27 pKa = 3.3VVNKK31 pKa = 9.97VEE33 pKa = 4.12QKK35 pKa = 9.21TLADD39 pKa = 3.33ILLQAFSSVLYY50 pKa = 10.76LGGLGFGTGKK60 pKa = 10.81GSTGTVGVRR69 pKa = 11.84PLPEE73 pKa = 3.97VPPIRR78 pKa = 11.84IPTEE82 pKa = 3.95TVPEE86 pKa = 4.8EE87 pKa = 4.16IPLAPIKK94 pKa = 10.17PKK96 pKa = 10.46TPITVGQGRR105 pKa = 11.84RR106 pKa = 11.84PFSVPLDD113 pKa = 3.73TIGAGFRR120 pKa = 11.84PVDD123 pKa = 3.55PSGSRR128 pKa = 11.84TIDD131 pKa = 3.44VIDD134 pKa = 4.32PSSPSVITPRR144 pKa = 11.84EE145 pKa = 4.12SLPDD149 pKa = 3.72TIITIGQPNLEE160 pKa = 4.37TGDD163 pKa = 4.1SVVTDD168 pKa = 4.23LNIITDD174 pKa = 3.69TTSIISHH181 pKa = 5.72PTVIQGVEE189 pKa = 4.06EE190 pKa = 4.01NVAILTVTPADD201 pKa = 3.9PPPTRR206 pKa = 11.84VKK208 pKa = 9.98FTLPEE213 pKa = 3.72QSPNISIEE221 pKa = 3.98SLAGHH226 pKa = 6.08VEE228 pKa = 3.62PTYY231 pKa = 11.22DD232 pKa = 3.58VFVDD236 pKa = 4.92PVASGEE242 pKa = 4.15DD243 pKa = 3.29IYY245 pKa = 11.21FGEE248 pKa = 5.51DD249 pKa = 3.12IPLEE253 pKa = 4.47PINPRR258 pKa = 11.84LEE260 pKa = 4.2FEE262 pKa = 4.29IEE264 pKa = 4.12EE265 pKa = 4.79PPQSSTPEE273 pKa = 3.62QRR275 pKa = 11.84LQRR278 pKa = 11.84AFSRR282 pKa = 11.84ARR284 pKa = 11.84EE285 pKa = 4.16FYY287 pKa = 10.38SRR289 pKa = 11.84HH290 pKa = 4.52IQQVRR295 pKa = 11.84TRR297 pKa = 11.84NLNLLGDD304 pKa = 3.82VARR307 pKa = 11.84AIEE310 pKa = 4.38FGFEE314 pKa = 3.7NPAFDD319 pKa = 5.25SEE321 pKa = 4.22ITLEE325 pKa = 4.32FEE327 pKa = 5.23RR328 pKa = 11.84EE329 pKa = 4.24VNDD332 pKa = 4.25LRR334 pKa = 11.84AAPDD338 pKa = 3.12IDD340 pKa = 3.7FTDD343 pKa = 3.14IQTISRR349 pKa = 11.84PYY351 pKa = 10.09IEE353 pKa = 4.77STPEE357 pKa = 3.37GTVRR361 pKa = 11.84VSRR364 pKa = 11.84LGTRR368 pKa = 11.84AGVRR372 pKa = 11.84TRR374 pKa = 11.84RR375 pKa = 11.84GTVLSQNVHH384 pKa = 5.72YY385 pKa = 10.16YY386 pKa = 10.85YY387 pKa = 10.43DD388 pKa = 3.64ISPIPSTEE396 pKa = 4.15TIEE399 pKa = 5.17LSTFTPEE406 pKa = 4.24TVVDD410 pKa = 3.6TSVNPNTEE418 pKa = 3.96SVFINEE424 pKa = 4.02IEE426 pKa = 4.13EE427 pKa = 4.31LPEE430 pKa = 4.91DD431 pKa = 4.0VLLDD435 pKa = 3.68SYY437 pKa = 11.25TEE439 pKa = 4.25SFNDD443 pKa = 3.17AHH445 pKa = 6.98LLLEE449 pKa = 4.55ATDD452 pKa = 4.19EE453 pKa = 4.14EE454 pKa = 5.16GEE456 pKa = 4.28RR457 pKa = 11.84YY458 pKa = 9.57QIPVVASDD466 pKa = 3.61TVKK469 pKa = 10.65VFVYY473 pKa = 10.26DD474 pKa = 3.65YY475 pKa = 11.12GSDD478 pKa = 3.57IIVAEE483 pKa = 4.56PGSSEE488 pKa = 4.64SPFPVPNIPLTPLQPPANILIVSDD512 pKa = 4.71DD513 pKa = 4.32FYY515 pKa = 11.53LHH517 pKa = 7.37PSLQRR522 pKa = 11.84KK523 pKa = 9.12RR524 pKa = 11.84KK525 pKa = 9.4RR526 pKa = 11.84KK527 pKa = 9.54RR528 pKa = 11.84LDD530 pKa = 3.15YY531 pKa = 10.84FF532 pKa = 3.88
MM1 pKa = 7.18SVSRR5 pKa = 11.84NKK7 pKa = 10.32RR8 pKa = 11.84DD9 pKa = 3.62TVEE12 pKa = 4.29NLYY15 pKa = 10.59KK16 pKa = 10.31SCKK19 pKa = 10.05LGGDD23 pKa = 3.95CPPDD27 pKa = 3.3VVNKK31 pKa = 9.97VEE33 pKa = 4.12QKK35 pKa = 9.21TLADD39 pKa = 3.33ILLQAFSSVLYY50 pKa = 10.76LGGLGFGTGKK60 pKa = 10.81GSTGTVGVRR69 pKa = 11.84PLPEE73 pKa = 3.97VPPIRR78 pKa = 11.84IPTEE82 pKa = 3.95TVPEE86 pKa = 4.8EE87 pKa = 4.16IPLAPIKK94 pKa = 10.17PKK96 pKa = 10.46TPITVGQGRR105 pKa = 11.84RR106 pKa = 11.84PFSVPLDD113 pKa = 3.73TIGAGFRR120 pKa = 11.84PVDD123 pKa = 3.55PSGSRR128 pKa = 11.84TIDD131 pKa = 3.44VIDD134 pKa = 4.32PSSPSVITPRR144 pKa = 11.84EE145 pKa = 4.12SLPDD149 pKa = 3.72TIITIGQPNLEE160 pKa = 4.37TGDD163 pKa = 4.1SVVTDD168 pKa = 4.23LNIITDD174 pKa = 3.69TTSIISHH181 pKa = 5.72PTVIQGVEE189 pKa = 4.06EE190 pKa = 4.01NVAILTVTPADD201 pKa = 3.9PPPTRR206 pKa = 11.84VKK208 pKa = 9.98FTLPEE213 pKa = 3.72QSPNISIEE221 pKa = 3.98SLAGHH226 pKa = 6.08VEE228 pKa = 3.62PTYY231 pKa = 11.22DD232 pKa = 3.58VFVDD236 pKa = 4.92PVASGEE242 pKa = 4.15DD243 pKa = 3.29IYY245 pKa = 11.21FGEE248 pKa = 5.51DD249 pKa = 3.12IPLEE253 pKa = 4.47PINPRR258 pKa = 11.84LEE260 pKa = 4.2FEE262 pKa = 4.29IEE264 pKa = 4.12EE265 pKa = 4.79PPQSSTPEE273 pKa = 3.62QRR275 pKa = 11.84LQRR278 pKa = 11.84AFSRR282 pKa = 11.84ARR284 pKa = 11.84EE285 pKa = 4.16FYY287 pKa = 10.38SRR289 pKa = 11.84HH290 pKa = 4.52IQQVRR295 pKa = 11.84TRR297 pKa = 11.84NLNLLGDD304 pKa = 3.82VARR307 pKa = 11.84AIEE310 pKa = 4.38FGFEE314 pKa = 3.7NPAFDD319 pKa = 5.25SEE321 pKa = 4.22ITLEE325 pKa = 4.32FEE327 pKa = 5.23RR328 pKa = 11.84EE329 pKa = 4.24VNDD332 pKa = 4.25LRR334 pKa = 11.84AAPDD338 pKa = 3.12IDD340 pKa = 3.7FTDD343 pKa = 3.14IQTISRR349 pKa = 11.84PYY351 pKa = 10.09IEE353 pKa = 4.77STPEE357 pKa = 3.37GTVRR361 pKa = 11.84VSRR364 pKa = 11.84LGTRR368 pKa = 11.84AGVRR372 pKa = 11.84TRR374 pKa = 11.84RR375 pKa = 11.84GTVLSQNVHH384 pKa = 5.72YY385 pKa = 10.16YY386 pKa = 10.85YY387 pKa = 10.43DD388 pKa = 3.64ISPIPSTEE396 pKa = 4.15TIEE399 pKa = 5.17LSTFTPEE406 pKa = 4.24TVVDD410 pKa = 3.6TSVNPNTEE418 pKa = 3.96SVFINEE424 pKa = 4.02IEE426 pKa = 4.13EE427 pKa = 4.31LPEE430 pKa = 4.91DD431 pKa = 4.0VLLDD435 pKa = 3.68SYY437 pKa = 11.25TEE439 pKa = 4.25SFNDD443 pKa = 3.17AHH445 pKa = 6.98LLLEE449 pKa = 4.55ATDD452 pKa = 4.19EE453 pKa = 4.14EE454 pKa = 5.16GEE456 pKa = 4.28RR457 pKa = 11.84YY458 pKa = 9.57QIPVVASDD466 pKa = 3.61TVKK469 pKa = 10.65VFVYY473 pKa = 10.26DD474 pKa = 3.65YY475 pKa = 11.12GSDD478 pKa = 3.57IIVAEE483 pKa = 4.56PGSSEE488 pKa = 4.64SPFPVPNIPLTPLQPPANILIVSDD512 pKa = 4.71DD513 pKa = 4.32FYY515 pKa = 11.53LHH517 pKa = 7.37PSLQRR522 pKa = 11.84KK523 pKa = 9.12RR524 pKa = 11.84KK525 pKa = 9.4RR526 pKa = 11.84KK527 pKa = 9.54RR528 pKa = 11.84LDD530 pKa = 3.15YY531 pKa = 10.84FF532 pKa = 3.88
Molecular weight: 58.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALY4|A0A2D2ALY4_9PAPI Protein E7 OS=Gammapapillomavirus 18 OX=1513263 GN=E7 PE=3 SV=1
MM1 pKa = 7.01NQADD5 pKa = 3.91LTKK8 pKa = 10.64RR9 pKa = 11.84SDD11 pKa = 3.51ALQEE15 pKa = 3.97RR16 pKa = 11.84LMNLYY21 pKa = 10.39EE22 pKa = 4.71EE23 pKa = 5.11GAQTIDD29 pKa = 3.53AQIEE33 pKa = 3.79HH34 pKa = 6.52WKK36 pKa = 10.21IIKK39 pKa = 8.71QQYY42 pKa = 6.29ITYY45 pKa = 9.3YY46 pKa = 8.2YY47 pKa = 10.64ARR49 pKa = 11.84KK50 pKa = 9.88EE51 pKa = 4.16GFKK54 pKa = 10.93KK55 pKa = 10.59LGLQILPTYY64 pKa = 10.24VVSEE68 pKa = 4.28YY69 pKa = 10.22KK70 pKa = 10.6AKK72 pKa = 10.54EE73 pKa = 4.11AIQQILLLEE82 pKa = 4.25SLKK85 pKa = 10.7KK86 pKa = 10.49SQFGNEE92 pKa = 3.75PWTLSEE98 pKa = 4.22TSAEE102 pKa = 4.18LTLTPPRR109 pKa = 11.84YY110 pKa = 9.81SFKK113 pKa = 10.45KK114 pKa = 9.82DD115 pKa = 3.32PYY117 pKa = 11.1VVDD120 pKa = 3.09VWFDD124 pKa = 3.83NNPEE128 pKa = 3.84NSFPYY133 pKa = 9.96TNWNKK138 pKa = 9.98IYY140 pKa = 10.52YY141 pKa = 9.48QDD143 pKa = 4.16EE144 pKa = 4.12QEE146 pKa = 3.95HH147 pKa = 4.4WHH149 pKa = 5.7RR150 pKa = 11.84AEE152 pKa = 4.15GKK154 pKa = 9.59VDD156 pKa = 3.21INGLYY161 pKa = 10.3FVDD164 pKa = 3.89INGDD168 pKa = 3.04RR169 pKa = 11.84NYY171 pKa = 10.5FVVFAPDD178 pKa = 3.17VEE180 pKa = 4.83RR181 pKa = 11.84YY182 pKa = 7.51STTGLWTVNFKK193 pKa = 10.68NQTISSVDD201 pKa = 3.26SSQTRR206 pKa = 11.84VSDD209 pKa = 3.51ISPQGSVSSSRR220 pKa = 11.84DD221 pKa = 3.35TIQRR225 pKa = 11.84PTTTSSRR232 pKa = 11.84GSEE235 pKa = 4.06GEE237 pKa = 3.73EE238 pKa = 3.63RR239 pKa = 11.84SPHH242 pKa = 5.17STTPSSPDD250 pKa = 2.94LRR252 pKa = 11.84GGQRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84QRR261 pKa = 11.84EE262 pKa = 3.51SSPRR266 pKa = 11.84GRR268 pKa = 11.84SKK270 pKa = 10.59RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84EE274 pKa = 3.87EE275 pKa = 3.74EE276 pKa = 3.89ATSGVAPGEE285 pKa = 4.22VGRR288 pKa = 11.84GSVTVPRR295 pKa = 11.84RR296 pKa = 11.84GLGRR300 pKa = 11.84IEE302 pKa = 4.39RR303 pKa = 11.84LTEE306 pKa = 3.72EE307 pKa = 4.87ARR309 pKa = 11.84DD310 pKa = 3.64PPIIFVKK317 pKa = 10.83GGANNLKK324 pKa = 9.87CWRR327 pKa = 11.84NRR329 pKa = 11.84CQKK332 pKa = 8.97TNRR335 pKa = 11.84LYY337 pKa = 10.62TVMSTVFRR345 pKa = 11.84YY346 pKa = 9.48PGTEE350 pKa = 3.44LGHH353 pKa = 6.64RR354 pKa = 11.84MVIGFRR360 pKa = 11.84DD361 pKa = 3.71FDD363 pKa = 3.59QRR365 pKa = 11.84TLFLKK370 pKa = 10.03TITFPKK376 pKa = 9.84GCTYY380 pKa = 10.65SLGRR384 pKa = 11.84IDD386 pKa = 4.88SLL388 pKa = 4.34
MM1 pKa = 7.01NQADD5 pKa = 3.91LTKK8 pKa = 10.64RR9 pKa = 11.84SDD11 pKa = 3.51ALQEE15 pKa = 3.97RR16 pKa = 11.84LMNLYY21 pKa = 10.39EE22 pKa = 4.71EE23 pKa = 5.11GAQTIDD29 pKa = 3.53AQIEE33 pKa = 3.79HH34 pKa = 6.52WKK36 pKa = 10.21IIKK39 pKa = 8.71QQYY42 pKa = 6.29ITYY45 pKa = 9.3YY46 pKa = 8.2YY47 pKa = 10.64ARR49 pKa = 11.84KK50 pKa = 9.88EE51 pKa = 4.16GFKK54 pKa = 10.93KK55 pKa = 10.59LGLQILPTYY64 pKa = 10.24VVSEE68 pKa = 4.28YY69 pKa = 10.22KK70 pKa = 10.6AKK72 pKa = 10.54EE73 pKa = 4.11AIQQILLLEE82 pKa = 4.25SLKK85 pKa = 10.7KK86 pKa = 10.49SQFGNEE92 pKa = 3.75PWTLSEE98 pKa = 4.22TSAEE102 pKa = 4.18LTLTPPRR109 pKa = 11.84YY110 pKa = 9.81SFKK113 pKa = 10.45KK114 pKa = 9.82DD115 pKa = 3.32PYY117 pKa = 11.1VVDD120 pKa = 3.09VWFDD124 pKa = 3.83NNPEE128 pKa = 3.84NSFPYY133 pKa = 9.96TNWNKK138 pKa = 9.98IYY140 pKa = 10.52YY141 pKa = 9.48QDD143 pKa = 4.16EE144 pKa = 4.12QEE146 pKa = 3.95HH147 pKa = 4.4WHH149 pKa = 5.7RR150 pKa = 11.84AEE152 pKa = 4.15GKK154 pKa = 9.59VDD156 pKa = 3.21INGLYY161 pKa = 10.3FVDD164 pKa = 3.89INGDD168 pKa = 3.04RR169 pKa = 11.84NYY171 pKa = 10.5FVVFAPDD178 pKa = 3.17VEE180 pKa = 4.83RR181 pKa = 11.84YY182 pKa = 7.51STTGLWTVNFKK193 pKa = 10.68NQTISSVDD201 pKa = 3.26SSQTRR206 pKa = 11.84VSDD209 pKa = 3.51ISPQGSVSSSRR220 pKa = 11.84DD221 pKa = 3.35TIQRR225 pKa = 11.84PTTTSSRR232 pKa = 11.84GSEE235 pKa = 4.06GEE237 pKa = 3.73EE238 pKa = 3.63RR239 pKa = 11.84SPHH242 pKa = 5.17STTPSSPDD250 pKa = 2.94LRR252 pKa = 11.84GGQRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84QRR261 pKa = 11.84EE262 pKa = 3.51SSPRR266 pKa = 11.84GRR268 pKa = 11.84SKK270 pKa = 10.59RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84EE274 pKa = 3.87EE275 pKa = 3.74EE276 pKa = 3.89ATSGVAPGEE285 pKa = 4.22VGRR288 pKa = 11.84GSVTVPRR295 pKa = 11.84RR296 pKa = 11.84GLGRR300 pKa = 11.84IEE302 pKa = 4.39RR303 pKa = 11.84LTEE306 pKa = 3.72EE307 pKa = 4.87ARR309 pKa = 11.84DD310 pKa = 3.64PPIIFVKK317 pKa = 10.83GGANNLKK324 pKa = 9.87CWRR327 pKa = 11.84NRR329 pKa = 11.84CQKK332 pKa = 8.97TNRR335 pKa = 11.84LYY337 pKa = 10.62TVMSTVFRR345 pKa = 11.84YY346 pKa = 9.48PGTEE350 pKa = 3.44LGHH353 pKa = 6.64RR354 pKa = 11.84MVIGFRR360 pKa = 11.84DD361 pKa = 3.71FDD363 pKa = 3.59QRR365 pKa = 11.84TLFLKK370 pKa = 10.03TITFPKK376 pKa = 9.84GCTYY380 pKa = 10.65SLGRR384 pKa = 11.84IDD386 pKa = 4.88SLL388 pKa = 4.34
Molecular weight: 44.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2258 |
96 |
597 |
376.3 |
42.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.606 ± 0.367 | 2.834 ± 1.293 |
6.289 ± 0.432 | 6.82 ± 0.607 |
5.049 ± 0.62 | 5.27 ± 0.639 |
1.284 ± 0.134 | 5.624 ± 0.699 |
5.359 ± 0.985 | 8.902 ± 0.967 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.55 ± 0.451 | 5.226 ± 0.606 |
6.289 ± 1.327 | 4.429 ± 0.433 |
5.802 ± 0.936 | 6.776 ± 0.81 |
6.643 ± 0.725 | 6.289 ± 0.732 |
1.151 ± 0.375 | 3.809 ± 0.63 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
