
Grapevine fleck virus (isolate Italy/MT48) (GFkV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Tymoviridae; Maculavirus; Grapevine fleck virus
Average proteome isoelectric point is 8.39
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
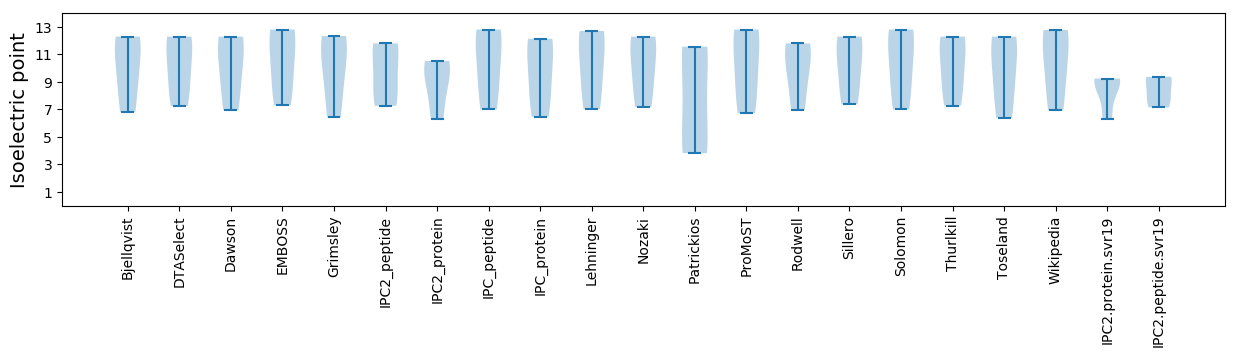
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8UZB6|RDRP_GFKVM RNA replication protein OS=Grapevine fleck virus (isolate Italy/MT48) OX=652668 PE=4 SV=1
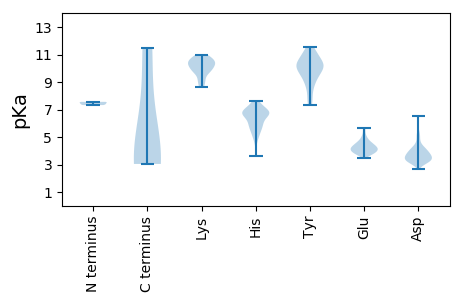
MM1 pKa = 7.56SLPADD6 pKa = 4.19LLLGAISSLLRR17 pKa = 11.84NPPTSDD23 pKa = 3.13AASPSADD30 pKa = 3.21QPAVSSSRR38 pKa = 11.84SDD40 pKa = 3.2SRR42 pKa = 11.84LVSAPLPAAPPAPTAIARR60 pKa = 11.84NPRR63 pKa = 11.84VSIHH67 pKa = 6.96LPFQFLWYY75 pKa = 9.71DD76 pKa = 3.15ITGTEE81 pKa = 3.7SSYY84 pKa = 11.12TSLSIASRR92 pKa = 11.84PEE94 pKa = 3.82VVTVARR100 pKa = 11.84PYY102 pKa = 9.56RR103 pKa = 11.84HH104 pKa = 6.52ARR106 pKa = 11.84LTSLEE111 pKa = 4.45AFVQPTASSATYY123 pKa = 8.56PQTVDD128 pKa = 3.54LCWTIDD134 pKa = 3.35SVTPARR140 pKa = 11.84SEE142 pKa = 3.81ILSVFGAQRR151 pKa = 11.84IAWGSVHH158 pKa = 6.78FSAPILLPAEE168 pKa = 4.43LSSLNPTIKK177 pKa = 10.73DD178 pKa = 3.34SVTYY182 pKa = 9.14TDD184 pKa = 5.3CPRR187 pKa = 11.84LTCGFYY193 pKa = 10.9RR194 pKa = 11.84NDD196 pKa = 3.09ACVALGSSAPICGSILIRR214 pKa = 11.84GVIEE218 pKa = 3.92CSAPINRR225 pKa = 11.84PTPSSS230 pKa = 3.3
MM1 pKa = 7.56SLPADD6 pKa = 4.19LLLGAISSLLRR17 pKa = 11.84NPPTSDD23 pKa = 3.13AASPSADD30 pKa = 3.21QPAVSSSRR38 pKa = 11.84SDD40 pKa = 3.2SRR42 pKa = 11.84LVSAPLPAAPPAPTAIARR60 pKa = 11.84NPRR63 pKa = 11.84VSIHH67 pKa = 6.96LPFQFLWYY75 pKa = 9.71DD76 pKa = 3.15ITGTEE81 pKa = 3.7SSYY84 pKa = 11.12TSLSIASRR92 pKa = 11.84PEE94 pKa = 3.82VVTVARR100 pKa = 11.84PYY102 pKa = 9.56RR103 pKa = 11.84HH104 pKa = 6.52ARR106 pKa = 11.84LTSLEE111 pKa = 4.45AFVQPTASSATYY123 pKa = 8.56PQTVDD128 pKa = 3.54LCWTIDD134 pKa = 3.35SVTPARR140 pKa = 11.84SEE142 pKa = 3.81ILSVFGAQRR151 pKa = 11.84IAWGSVHH158 pKa = 6.78FSAPILLPAEE168 pKa = 4.43LSSLNPTIKK177 pKa = 10.73DD178 pKa = 3.34SVTYY182 pKa = 9.14TDD184 pKa = 5.3CPRR187 pKa = 11.84LTCGFYY193 pKa = 10.9RR194 pKa = 11.84NDD196 pKa = 3.09ACVALGSSAPICGSILIRR214 pKa = 11.84GVIEE218 pKa = 3.92CSAPINRR225 pKa = 11.84PTPSSS230 pKa = 3.3
Molecular weight: 24.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8UZB4|ORF3_GFKVM Uncharacterized protein ORF3 OS=Grapevine fleck virus (isolate Italy/MT48) OX=652668 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.32NRR3 pKa = 11.84GPPLRR8 pKa = 11.84SRR10 pKa = 11.84PPSSPPPASAFPGPSPFPSPSPANSLPSASPPPPTCTPSSPVSRR54 pKa = 11.84PFASARR60 pKa = 11.84LRR62 pKa = 11.84TSHH65 pKa = 7.5PPRR68 pKa = 11.84CPHH71 pKa = 6.88RR72 pKa = 11.84SAPPSAPSPPFTPPHH87 pKa = 6.68PLPTPTPSSSPRR99 pKa = 11.84SPWLSLAPLPTSSASLASFPPPPSSFSSPSSPSTSPLSPSSSSFPSSSSFSFLVPSNSS157 pKa = 3.04
MM1 pKa = 7.32NRR3 pKa = 11.84GPPLRR8 pKa = 11.84SRR10 pKa = 11.84PPSSPPPASAFPGPSPFPSPSPANSLPSASPPPPTCTPSSPVSRR54 pKa = 11.84PFASARR60 pKa = 11.84LRR62 pKa = 11.84TSHH65 pKa = 7.5PPRR68 pKa = 11.84CPHH71 pKa = 6.88RR72 pKa = 11.84SAPPSAPSPPFTPPHH87 pKa = 6.68PLPTPTPSSSPRR99 pKa = 11.84SPWLSLAPLPTSSASLASFPPPPSSFSSPSSPSTSPLSPSSSSFPSSSSFSFLVPSNSS157 pKa = 3.04
Molecular weight: 15.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2645 |
157 |
1949 |
661.3 |
71.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.603 ± 0.614 | 1.172 ± 0.303 |
3.289 ± 0.925 | 2.457 ± 0.755 |
4.688 ± 0.884 | 3.138 ± 0.735 |
3.781 ± 1.136 | 3.516 ± 0.896 |
1.701 ± 0.667 | 12.703 ± 1.367 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.567 ± 0.03 | 2.155 ± 0.414 |
16.295 ± 5.454 | 2.533 ± 0.837 |
6.238 ± 0.222 | 10.888 ± 4.771 |
7.486 ± 0.808 | 4.423 ± 0.851 |
1.134 ± 0.327 | 2.231 ± 0.723 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
