
Firmicutes bacterium CAG:552
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
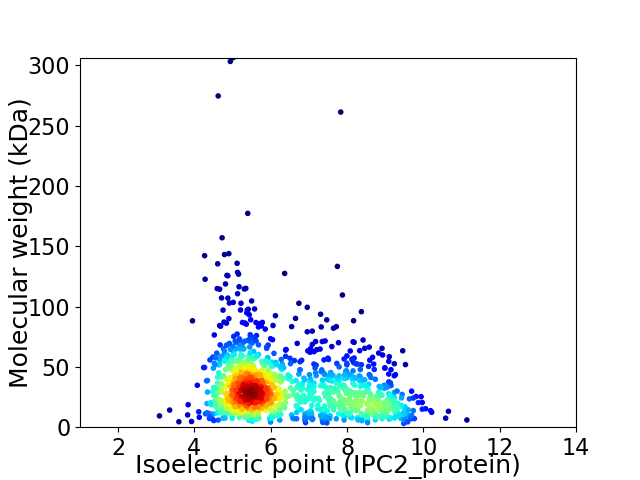
Virtual 2D-PAGE plot for 1175 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
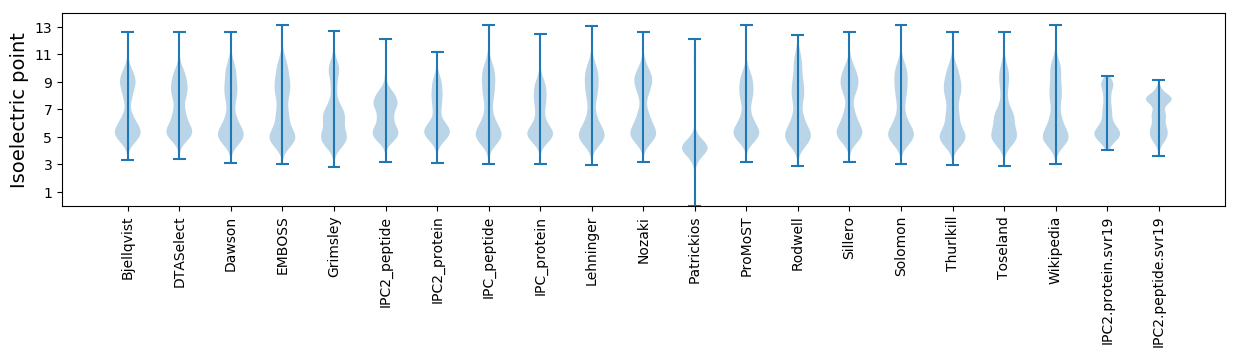
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6GWI6|R6GWI6_9FIRM Membrane protein insertase YidC OS=Firmicutes bacterium CAG:552 OX=1263029 GN=BN704_00677 PE=3 SV=1
MM1 pKa = 7.27MNGLSVLPQNEE12 pKa = 4.39NLTVPAFTEE21 pKa = 3.95LLEE24 pKa = 6.21DD25 pKa = 2.92IFGYY29 pKa = 10.6LKK31 pKa = 10.85GYY33 pKa = 6.37MHH35 pKa = 7.08CKK37 pKa = 9.44IAFICDD43 pKa = 3.16TDD45 pKa = 3.41EE46 pKa = 4.9SKK48 pKa = 10.89FAGYY52 pKa = 8.29ATFLNYY58 pKa = 10.42LDD60 pKa = 3.52IHH62 pKa = 6.94VNTDD66 pKa = 2.02IRR68 pKa = 11.84TPFAHH73 pKa = 5.69SVMYY77 pKa = 10.38RR78 pKa = 11.84ISPLIEE84 pKa = 4.47ANTPNDD90 pKa = 3.9MEE92 pKa = 4.46IMILFDD98 pKa = 3.07SHH100 pKa = 7.21YY101 pKa = 11.39ADD103 pKa = 5.34AYY105 pKa = 11.47NDD107 pKa = 3.7AQNADD112 pKa = 3.08VTYY115 pKa = 10.57NLNDD119 pKa = 3.47RR120 pKa = 11.84TSYY123 pKa = 11.12QIMSEE128 pKa = 3.53IMYY131 pKa = 10.35VLEE134 pKa = 4.87DD135 pKa = 3.39MCNAQSSSLNDD146 pKa = 2.94IAQACRR152 pKa = 11.84VNYY155 pKa = 9.71NVRR158 pKa = 11.84LYY160 pKa = 10.52SVSPGGIFKK169 pKa = 10.69NLEE172 pKa = 3.89NYY174 pKa = 7.28TTISMTDD181 pKa = 2.7AVYY184 pKa = 10.19NAKK187 pKa = 10.07EE188 pKa = 3.79LVYY191 pKa = 10.78IMRR194 pKa = 11.84PDD196 pKa = 3.34ANLNLNCRR204 pKa = 11.84LLEE207 pKa = 4.16DD208 pKa = 5.46LICSFTPDD216 pKa = 3.29DD217 pKa = 4.22FYY219 pKa = 11.55AYY221 pKa = 10.46SFGSVSVPAPEE232 pKa = 4.85ASLLTIYY239 pKa = 10.5EE240 pKa = 3.92LVYY243 pKa = 10.2TYY245 pKa = 9.47EE246 pKa = 3.9EE247 pKa = 4.9CFTRR251 pKa = 11.84RR252 pKa = 11.84VEE254 pKa = 4.16AMIDD258 pKa = 3.64FLEE261 pKa = 6.04DD262 pKa = 3.55EE263 pKa = 5.22DD264 pKa = 6.02LSDD267 pKa = 3.53YY268 pKa = 10.92NNWYY272 pKa = 9.28GVCDD276 pKa = 3.67ISYY279 pKa = 9.92KK280 pKa = 10.45ILGTGPDD287 pKa = 2.92GWMISVLGYY296 pKa = 9.99GSKK299 pKa = 10.5VKK301 pKa = 10.09PLYY304 pKa = 10.45DD305 pKa = 3.18
MM1 pKa = 7.27MNGLSVLPQNEE12 pKa = 4.39NLTVPAFTEE21 pKa = 3.95LLEE24 pKa = 6.21DD25 pKa = 2.92IFGYY29 pKa = 10.6LKK31 pKa = 10.85GYY33 pKa = 6.37MHH35 pKa = 7.08CKK37 pKa = 9.44IAFICDD43 pKa = 3.16TDD45 pKa = 3.41EE46 pKa = 4.9SKK48 pKa = 10.89FAGYY52 pKa = 8.29ATFLNYY58 pKa = 10.42LDD60 pKa = 3.52IHH62 pKa = 6.94VNTDD66 pKa = 2.02IRR68 pKa = 11.84TPFAHH73 pKa = 5.69SVMYY77 pKa = 10.38RR78 pKa = 11.84ISPLIEE84 pKa = 4.47ANTPNDD90 pKa = 3.9MEE92 pKa = 4.46IMILFDD98 pKa = 3.07SHH100 pKa = 7.21YY101 pKa = 11.39ADD103 pKa = 5.34AYY105 pKa = 11.47NDD107 pKa = 3.7AQNADD112 pKa = 3.08VTYY115 pKa = 10.57NLNDD119 pKa = 3.47RR120 pKa = 11.84TSYY123 pKa = 11.12QIMSEE128 pKa = 3.53IMYY131 pKa = 10.35VLEE134 pKa = 4.87DD135 pKa = 3.39MCNAQSSSLNDD146 pKa = 2.94IAQACRR152 pKa = 11.84VNYY155 pKa = 9.71NVRR158 pKa = 11.84LYY160 pKa = 10.52SVSPGGIFKK169 pKa = 10.69NLEE172 pKa = 3.89NYY174 pKa = 7.28TTISMTDD181 pKa = 2.7AVYY184 pKa = 10.19NAKK187 pKa = 10.07EE188 pKa = 3.79LVYY191 pKa = 10.78IMRR194 pKa = 11.84PDD196 pKa = 3.34ANLNLNCRR204 pKa = 11.84LLEE207 pKa = 4.16DD208 pKa = 5.46LICSFTPDD216 pKa = 3.29DD217 pKa = 4.22FYY219 pKa = 11.55AYY221 pKa = 10.46SFGSVSVPAPEE232 pKa = 4.85ASLLTIYY239 pKa = 10.5EE240 pKa = 3.92LVYY243 pKa = 10.2TYY245 pKa = 9.47EE246 pKa = 3.9EE247 pKa = 4.9CFTRR251 pKa = 11.84RR252 pKa = 11.84VEE254 pKa = 4.16AMIDD258 pKa = 3.64FLEE261 pKa = 6.04DD262 pKa = 3.55EE263 pKa = 5.22DD264 pKa = 6.02LSDD267 pKa = 3.53YY268 pKa = 10.92NNWYY272 pKa = 9.28GVCDD276 pKa = 3.67ISYY279 pKa = 9.92KK280 pKa = 10.45ILGTGPDD287 pKa = 2.92GWMISVLGYY296 pKa = 9.99GSKK299 pKa = 10.5VKK301 pKa = 10.09PLYY304 pKa = 10.45DD305 pKa = 3.18
Molecular weight: 34.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6GXY9|R6GXY9_9FIRM Phosphomannomutase OS=Firmicutes bacterium CAG:552 OX=1263029 GN=BN704_00880 PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.7QPKK8 pKa = 9.37KK9 pKa = 8.53RR10 pKa = 11.84ARR12 pKa = 11.84QKK14 pKa = 9.77VHH16 pKa = 6.35GFRR19 pKa = 11.84QRR21 pKa = 11.84MKK23 pKa = 7.98TQGGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.15GRR39 pKa = 11.84KK40 pKa = 8.25NISVKK45 pKa = 10.49PLGSHH50 pKa = 6.88
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.7QPKK8 pKa = 9.37KK9 pKa = 8.53RR10 pKa = 11.84ARR12 pKa = 11.84QKK14 pKa = 9.77VHH16 pKa = 6.35GFRR19 pKa = 11.84QRR21 pKa = 11.84MKK23 pKa = 7.98TQGGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.15GRR39 pKa = 11.84KK40 pKa = 8.25NISVKK45 pKa = 10.49PLGSHH50 pKa = 6.88
Molecular weight: 6.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
378033 |
30 |
2861 |
321.7 |
35.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.987 ± 0.061 | 1.808 ± 0.036 |
5.97 ± 0.066 | 6.548 ± 0.071 |
4.372 ± 0.052 | 7.113 ± 0.06 |
1.277 ± 0.026 | 7.609 ± 0.055 |
7.66 ± 0.061 | 8.629 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.037 | 4.504 ± 0.053 |
3.015 ± 0.03 | 2.285 ± 0.031 |
4.136 ± 0.06 | 6.693 ± 0.07 |
5.428 ± 0.076 | 7.802 ± 0.068 |
0.565 ± 0.019 | 4.053 ± 0.057 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
