
Lake Sarah-associated circular molecule 12
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQP3|A0A140AQP3_9VIRU Uncharacterized protein OS=Lake Sarah-associated circular molecule 12 OX=1809112 PE=4 SV=1
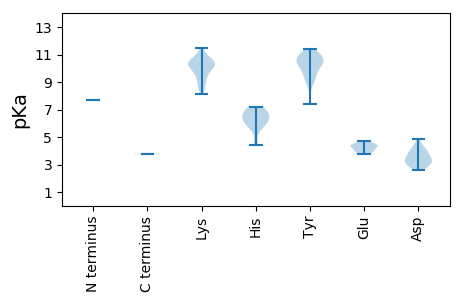
MM1 pKa = 7.66PGVKK5 pKa = 10.13EE6 pKa = 3.82SDD8 pKa = 2.63KK9 pKa = 10.5WFIRR13 pKa = 11.84ITAPHH18 pKa = 6.22TLLRR22 pKa = 11.84PKK24 pKa = 10.43VEE26 pKa = 4.03EE27 pKa = 4.26LKK29 pKa = 11.04GQIDD33 pKa = 4.18IKK35 pKa = 10.93RR36 pKa = 11.84MAIGFHH42 pKa = 5.96IGNRR46 pKa = 11.84TGKK49 pKa = 8.51EE50 pKa = 3.97HH51 pKa = 6.3IHH53 pKa = 6.49IALEE57 pKa = 4.02LLGNLQQQSINKK69 pKa = 9.06RR70 pKa = 11.84MKK72 pKa = 10.43KK73 pKa = 9.58IFGVSGADD81 pKa = 3.38YY82 pKa = 10.8SSKK85 pKa = 10.09IWDD88 pKa = 3.93GSHH91 pKa = 6.76KK92 pKa = 10.23VLSYY96 pKa = 10.67LYY98 pKa = 9.23HH99 pKa = 6.91DD100 pKa = 4.13EE101 pKa = 4.5KK102 pKa = 11.48GSVEE106 pKa = 4.09YY107 pKa = 10.4FKK109 pKa = 10.9MEE111 pKa = 4.21LTPSEE116 pKa = 4.7LEE118 pKa = 4.29EE119 pKa = 3.93IMKK122 pKa = 9.9TKK124 pKa = 10.45DD125 pKa = 3.1VYY127 pKa = 11.4KK128 pKa = 10.61DD129 pKa = 3.12IVHH132 pKa = 6.29TAKK135 pKa = 10.44QKK137 pKa = 9.74AATRR141 pKa = 11.84IPEE144 pKa = 4.68RR145 pKa = 11.84ILEE148 pKa = 4.44EE149 pKa = 3.8IRR151 pKa = 11.84EE152 pKa = 4.33SGSTWTMRR160 pKa = 11.84QIIKK164 pKa = 10.47RR165 pKa = 11.84IFVGVNKK172 pKa = 10.35DD173 pKa = 2.62EE174 pKa = 4.38WYY176 pKa = 9.91PPGSQMEE183 pKa = 4.37RR184 pKa = 11.84YY185 pKa = 8.78VQEE188 pKa = 4.11ILVKK192 pKa = 10.37QNPEE196 pKa = 3.75DD197 pKa = 4.3SIPALTDD204 pKa = 3.1YY205 pKa = 11.14YY206 pKa = 10.92LARR209 pKa = 11.84FTYY212 pKa = 10.63GLL214 pKa = 3.75
MM1 pKa = 7.66PGVKK5 pKa = 10.13EE6 pKa = 3.82SDD8 pKa = 2.63KK9 pKa = 10.5WFIRR13 pKa = 11.84ITAPHH18 pKa = 6.22TLLRR22 pKa = 11.84PKK24 pKa = 10.43VEE26 pKa = 4.03EE27 pKa = 4.26LKK29 pKa = 11.04GQIDD33 pKa = 4.18IKK35 pKa = 10.93RR36 pKa = 11.84MAIGFHH42 pKa = 5.96IGNRR46 pKa = 11.84TGKK49 pKa = 8.51EE50 pKa = 3.97HH51 pKa = 6.3IHH53 pKa = 6.49IALEE57 pKa = 4.02LLGNLQQQSINKK69 pKa = 9.06RR70 pKa = 11.84MKK72 pKa = 10.43KK73 pKa = 9.58IFGVSGADD81 pKa = 3.38YY82 pKa = 10.8SSKK85 pKa = 10.09IWDD88 pKa = 3.93GSHH91 pKa = 6.76KK92 pKa = 10.23VLSYY96 pKa = 10.67LYY98 pKa = 9.23HH99 pKa = 6.91DD100 pKa = 4.13EE101 pKa = 4.5KK102 pKa = 11.48GSVEE106 pKa = 4.09YY107 pKa = 10.4FKK109 pKa = 10.9MEE111 pKa = 4.21LTPSEE116 pKa = 4.7LEE118 pKa = 4.29EE119 pKa = 3.93IMKK122 pKa = 9.9TKK124 pKa = 10.45DD125 pKa = 3.1VYY127 pKa = 11.4KK128 pKa = 10.61DD129 pKa = 3.12IVHH132 pKa = 6.29TAKK135 pKa = 10.44QKK137 pKa = 9.74AATRR141 pKa = 11.84IPEE144 pKa = 4.68RR145 pKa = 11.84ILEE148 pKa = 4.44EE149 pKa = 3.8IRR151 pKa = 11.84EE152 pKa = 4.33SGSTWTMRR160 pKa = 11.84QIIKK164 pKa = 10.47RR165 pKa = 11.84IFVGVNKK172 pKa = 10.35DD173 pKa = 2.62EE174 pKa = 4.38WYY176 pKa = 9.91PPGSQMEE183 pKa = 4.37RR184 pKa = 11.84YY185 pKa = 8.78VQEE188 pKa = 4.11ILVKK192 pKa = 10.37QNPEE196 pKa = 3.75DD197 pKa = 4.3SIPALTDD204 pKa = 3.1YY205 pKa = 11.14YY206 pKa = 10.92LARR209 pKa = 11.84FTYY212 pKa = 10.63GLL214 pKa = 3.75
Molecular weight: 24.88 kDa
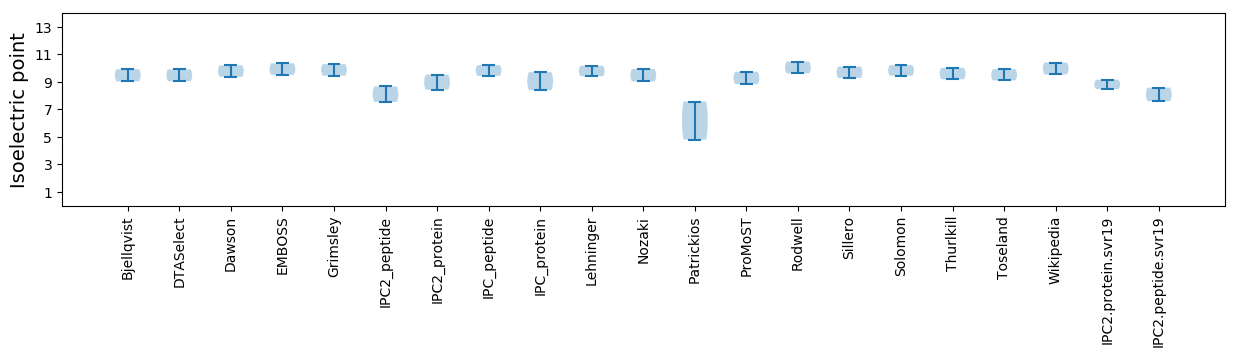
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQP3|A0A140AQP3_9VIRU Uncharacterized protein OS=Lake Sarah-associated circular molecule 12 OX=1809112 PE=4 SV=1
MM1 pKa = 7.66PPGRR5 pKa = 11.84KK6 pKa = 8.66YY7 pKa = 10.33RR8 pKa = 11.84KK9 pKa = 9.49KK10 pKa = 9.13GSKK13 pKa = 9.1PKK15 pKa = 9.98RR16 pKa = 11.84VLMPYY21 pKa = 9.45TPARR25 pKa = 11.84KK26 pKa = 8.48YY27 pKa = 10.18KK28 pKa = 10.24KK29 pKa = 9.86RR30 pKa = 11.84GGRR33 pKa = 11.84STAGRR38 pKa = 11.84LITTMAGLLTNSTWTQKK55 pKa = 10.1ARR57 pKa = 11.84KK58 pKa = 9.22LGFQARR64 pKa = 11.84AMRR67 pKa = 11.84AVGAPNIYY75 pKa = 9.57HH76 pKa = 7.19ANFALPVVVPPGLQDD91 pKa = 4.88FISFPTLQRR100 pKa = 11.84TQLQAILSSVPQSSGLNRR118 pKa = 11.84ALIQSAQTEE127 pKa = 4.44LTFTNVTNAPVQVEE141 pKa = 4.48MYY143 pKa = 10.47DD144 pKa = 2.94IMFRR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.35LSTDD154 pKa = 3.04ALVVNNGNNFTFATIEE170 pKa = 3.91EE171 pKa = 4.58MIKK174 pKa = 10.46SGCQAANGIAPGGTDD189 pKa = 3.03VSSYY193 pKa = 10.43IGASAYY199 pKa = 10.32DD200 pKa = 3.47SQIFKK205 pKa = 10.41DD206 pKa = 3.4YY207 pKa = 10.78CRR209 pKa = 11.84VVKK212 pKa = 10.27RR213 pKa = 11.84QHH215 pKa = 5.66VLLASGATHH224 pKa = 7.0RR225 pKa = 11.84HH226 pKa = 4.45QSNIGINKK234 pKa = 9.73IIDD237 pKa = 3.59EE238 pKa = 4.38SVGANEE244 pKa = 4.26DD245 pKa = 3.8LIYY248 pKa = 11.21VKK250 pKa = 10.61GYY252 pKa = 9.25TYY254 pKa = 10.4ATLILTRR261 pKa = 11.84GVGAFDD267 pKa = 3.59ATASVPNSTTNGAFLNFVTSVRR289 pKa = 11.84IKK291 pKa = 8.11YY292 pKa = 7.4TWVADD297 pKa = 3.91TTNSVYY303 pKa = 10.83YY304 pKa = 10.6HH305 pKa = 6.68NLPLHH310 pKa = 5.87TGVPNVRR317 pKa = 11.84NTGSGAYY324 pKa = 9.56EE325 pKa = 4.2SEE327 pKa = 4.55TPP329 pKa = 3.8
MM1 pKa = 7.66PPGRR5 pKa = 11.84KK6 pKa = 8.66YY7 pKa = 10.33RR8 pKa = 11.84KK9 pKa = 9.49KK10 pKa = 9.13GSKK13 pKa = 9.1PKK15 pKa = 9.98RR16 pKa = 11.84VLMPYY21 pKa = 9.45TPARR25 pKa = 11.84KK26 pKa = 8.48YY27 pKa = 10.18KK28 pKa = 10.24KK29 pKa = 9.86RR30 pKa = 11.84GGRR33 pKa = 11.84STAGRR38 pKa = 11.84LITTMAGLLTNSTWTQKK55 pKa = 10.1ARR57 pKa = 11.84KK58 pKa = 9.22LGFQARR64 pKa = 11.84AMRR67 pKa = 11.84AVGAPNIYY75 pKa = 9.57HH76 pKa = 7.19ANFALPVVVPPGLQDD91 pKa = 4.88FISFPTLQRR100 pKa = 11.84TQLQAILSSVPQSSGLNRR118 pKa = 11.84ALIQSAQTEE127 pKa = 4.44LTFTNVTNAPVQVEE141 pKa = 4.48MYY143 pKa = 10.47DD144 pKa = 2.94IMFRR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.35LSTDD154 pKa = 3.04ALVVNNGNNFTFATIEE170 pKa = 3.91EE171 pKa = 4.58MIKK174 pKa = 10.46SGCQAANGIAPGGTDD189 pKa = 3.03VSSYY193 pKa = 10.43IGASAYY199 pKa = 10.32DD200 pKa = 3.47SQIFKK205 pKa = 10.41DD206 pKa = 3.4YY207 pKa = 10.78CRR209 pKa = 11.84VVKK212 pKa = 10.27RR213 pKa = 11.84QHH215 pKa = 5.66VLLASGATHH224 pKa = 7.0RR225 pKa = 11.84HH226 pKa = 4.45QSNIGINKK234 pKa = 9.73IIDD237 pKa = 3.59EE238 pKa = 4.38SVGANEE244 pKa = 4.26DD245 pKa = 3.8LIYY248 pKa = 11.21VKK250 pKa = 10.61GYY252 pKa = 9.25TYY254 pKa = 10.4ATLILTRR261 pKa = 11.84GVGAFDD267 pKa = 3.59ATASVPNSTTNGAFLNFVTSVRR289 pKa = 11.84IKK291 pKa = 8.11YY292 pKa = 7.4TWVADD297 pKa = 3.91TTNSVYY303 pKa = 10.83YY304 pKa = 10.6HH305 pKa = 6.68NLPLHH310 pKa = 5.87TGVPNVRR317 pKa = 11.84NTGSGAYY324 pKa = 9.56EE325 pKa = 4.2SEE327 pKa = 4.55TPP329 pKa = 3.8
Molecular weight: 35.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
543 |
214 |
329 |
271.5 |
30.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.366 ± 1.919 | 0.368 ± 0.224 |
3.867 ± 0.489 | 4.972 ± 2.371 |
3.315 ± 0.31 | 7.366 ± 0.5 |
2.394 ± 0.532 | 7.182 ± 1.597 |
6.814 ± 1.821 | 7.182 ± 0.179 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.578 ± 0.421 | 4.788 ± 1.488 |
4.788 ± 0.354 | 4.236 ± 0.018 |
5.893 ± 0.173 | 6.814 ± 0.449 |
7.735 ± 1.575 | 6.63 ± 0.904 |
1.105 ± 0.464 | 4.604 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
