
Pea enation mosaic virus-2 (PEMV-2)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus
Average proteome isoelectric point is 7.97
Get precalculated fractions of proteins
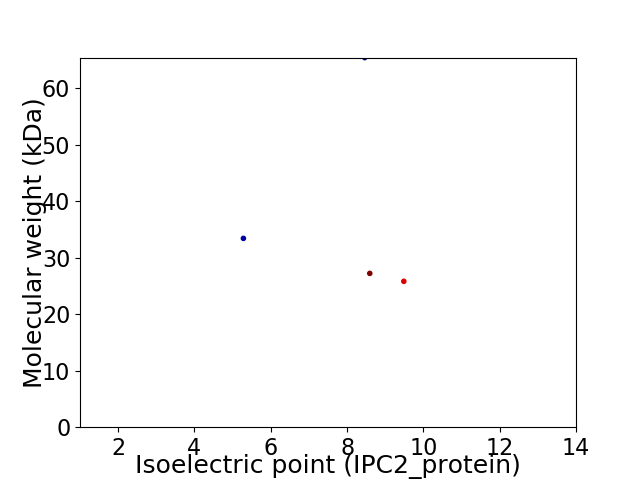
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q84694|Q84694_PEMV2 RNA-directed RNA polymerase (Fragment) OS=Pea enation mosaic virus-2 OX=193120 PE=2 SV=1
MM1 pKa = 7.2NCVARR6 pKa = 11.84IKK8 pKa = 10.5RR9 pKa = 11.84WFTPDD14 pKa = 3.54FTPGEE19 pKa = 4.44GVKK22 pKa = 10.42SRR24 pKa = 11.84AQLEE28 pKa = 4.46RR29 pKa = 11.84EE30 pKa = 4.44LDD32 pKa = 3.76PTWALLVCQEE42 pKa = 4.1RR43 pKa = 11.84ARR45 pKa = 11.84RR46 pKa = 11.84DD47 pKa = 3.18ADD49 pKa = 3.79SIANEE54 pKa = 4.02WYY56 pKa = 9.57EE57 pKa = 4.82GSMEE61 pKa = 4.51CNLLIPRR68 pKa = 11.84PTTEE72 pKa = 4.82DD73 pKa = 3.32VFGPSIAPEE82 pKa = 3.73PVALVEE88 pKa = 3.89EE89 pKa = 4.92TTRR92 pKa = 11.84SRR94 pKa = 11.84APCVDD99 pKa = 3.27VPAEE103 pKa = 4.13EE104 pKa = 4.44SCKK107 pKa = 10.45SAEE110 pKa = 4.1IDD112 pKa = 3.7PVDD115 pKa = 3.64LAKK118 pKa = 10.36FDD120 pKa = 3.88SLHH123 pKa = 6.36RR124 pKa = 11.84RR125 pKa = 11.84LLAEE129 pKa = 4.48ANPCRR134 pKa = 11.84EE135 pKa = 4.0MVLWVPPGLPAEE147 pKa = 4.22RR148 pKa = 11.84DD149 pKa = 3.54VLPRR153 pKa = 11.84ARR155 pKa = 11.84GVIMIPEE162 pKa = 4.43VPASAHH168 pKa = 4.81TLSVKK173 pKa = 10.31VMEE176 pKa = 4.68AVRR179 pKa = 11.84LAQEE183 pKa = 4.31VLASLAKK190 pKa = 9.94RR191 pKa = 11.84ALEE194 pKa = 4.01KK195 pKa = 10.66RR196 pKa = 11.84STPTLTAQAQPEE208 pKa = 4.27ATLSGCDD215 pKa = 3.33YY216 pKa = 10.53PYY218 pKa = 11.0QEE220 pKa = 4.71TGAAAAWITPGCIAMEE236 pKa = 4.24LRR238 pKa = 11.84AKK240 pKa = 10.67FGVCKK245 pKa = 9.71RR246 pKa = 11.84TPANLEE252 pKa = 3.58MGSRR256 pKa = 11.84VARR259 pKa = 11.84EE260 pKa = 4.06LLRR263 pKa = 11.84DD264 pKa = 3.42NCVTCRR270 pKa = 11.84EE271 pKa = 4.47TTWYY275 pKa = 9.21TSAIAVDD282 pKa = 4.23LWLTPTVVDD291 pKa = 4.33LACGRR296 pKa = 11.84RR297 pKa = 11.84AADD300 pKa = 3.98FWW302 pKa = 5.89
MM1 pKa = 7.2NCVARR6 pKa = 11.84IKK8 pKa = 10.5RR9 pKa = 11.84WFTPDD14 pKa = 3.54FTPGEE19 pKa = 4.44GVKK22 pKa = 10.42SRR24 pKa = 11.84AQLEE28 pKa = 4.46RR29 pKa = 11.84EE30 pKa = 4.44LDD32 pKa = 3.76PTWALLVCQEE42 pKa = 4.1RR43 pKa = 11.84ARR45 pKa = 11.84RR46 pKa = 11.84DD47 pKa = 3.18ADD49 pKa = 3.79SIANEE54 pKa = 4.02WYY56 pKa = 9.57EE57 pKa = 4.82GSMEE61 pKa = 4.51CNLLIPRR68 pKa = 11.84PTTEE72 pKa = 4.82DD73 pKa = 3.32VFGPSIAPEE82 pKa = 3.73PVALVEE88 pKa = 3.89EE89 pKa = 4.92TTRR92 pKa = 11.84SRR94 pKa = 11.84APCVDD99 pKa = 3.27VPAEE103 pKa = 4.13EE104 pKa = 4.44SCKK107 pKa = 10.45SAEE110 pKa = 4.1IDD112 pKa = 3.7PVDD115 pKa = 3.64LAKK118 pKa = 10.36FDD120 pKa = 3.88SLHH123 pKa = 6.36RR124 pKa = 11.84RR125 pKa = 11.84LLAEE129 pKa = 4.48ANPCRR134 pKa = 11.84EE135 pKa = 4.0MVLWVPPGLPAEE147 pKa = 4.22RR148 pKa = 11.84DD149 pKa = 3.54VLPRR153 pKa = 11.84ARR155 pKa = 11.84GVIMIPEE162 pKa = 4.43VPASAHH168 pKa = 4.81TLSVKK173 pKa = 10.31VMEE176 pKa = 4.68AVRR179 pKa = 11.84LAQEE183 pKa = 4.31VLASLAKK190 pKa = 9.94RR191 pKa = 11.84ALEE194 pKa = 4.01KK195 pKa = 10.66RR196 pKa = 11.84STPTLTAQAQPEE208 pKa = 4.27ATLSGCDD215 pKa = 3.33YY216 pKa = 10.53PYY218 pKa = 11.0QEE220 pKa = 4.71TGAAAAWITPGCIAMEE236 pKa = 4.24LRR238 pKa = 11.84AKK240 pKa = 10.67FGVCKK245 pKa = 9.71RR246 pKa = 11.84TPANLEE252 pKa = 3.58MGSRR256 pKa = 11.84VARR259 pKa = 11.84EE260 pKa = 4.06LLRR263 pKa = 11.84DD264 pKa = 3.42NCVTCRR270 pKa = 11.84EE271 pKa = 4.47TTWYY275 pKa = 9.21TSAIAVDD282 pKa = 4.23LWLTPTVVDD291 pKa = 4.33LACGRR296 pKa = 11.84RR297 pKa = 11.84AADD300 pKa = 3.98FWW302 pKa = 5.89
Molecular weight: 33.41 kDa
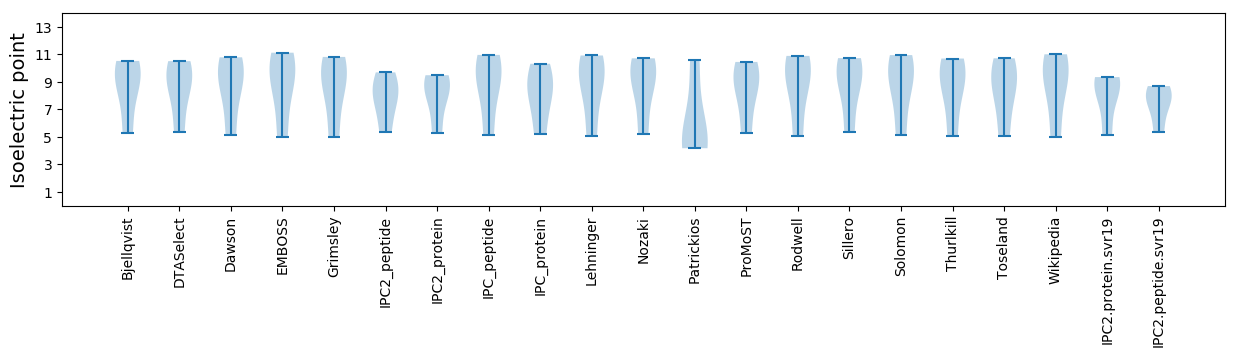
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84696|Q84696_PEMV2 27kDa protein OS=Pea enation mosaic virus-2 OX=193120 PE=2 SV=1
MM1 pKa = 7.66AVGKK5 pKa = 10.66YY6 pKa = 6.42MTIIINVNNDD16 pKa = 2.65EE17 pKa = 4.23RR18 pKa = 11.84KK19 pKa = 9.36QPEE22 pKa = 4.32GATGSSVRR30 pKa = 11.84RR31 pKa = 11.84GDD33 pKa = 3.53NKK35 pKa = 9.44RR36 pKa = 11.84TRR38 pKa = 11.84GNKK41 pKa = 8.59PRR43 pKa = 11.84SHH45 pKa = 6.71HH46 pKa = 5.96PGSRR50 pKa = 11.84EE51 pKa = 3.51RR52 pKa = 11.84KK53 pKa = 8.9GYY55 pKa = 9.09NHH57 pKa = 7.64PSPTPKK63 pKa = 10.09NSKK66 pKa = 9.6QGQLRR71 pKa = 11.84TEE73 pKa = 4.48AVQEE77 pKa = 4.25HH78 pKa = 7.11PKK80 pKa = 10.66HH81 pKa = 6.2GGTAFRR87 pKa = 11.84RR88 pKa = 11.84EE89 pKa = 4.5SGGSVHH95 pKa = 7.01PSHH98 pKa = 7.17PRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84ARR104 pKa = 11.84RR105 pKa = 11.84GGDD108 pKa = 3.06MAPRR112 pKa = 11.84QHH114 pKa = 6.96PPPPRR119 pKa = 11.84EE120 pKa = 3.54RR121 pKa = 11.84RR122 pKa = 11.84TKK124 pKa = 10.5TEE126 pKa = 3.69TQAEE130 pKa = 4.04RR131 pKa = 11.84RR132 pKa = 11.84AQALSVLPTLLDD144 pKa = 4.38CIGGLDD150 pKa = 3.94LGPAEE155 pKa = 4.56VLLHH159 pKa = 5.4CHH161 pKa = 5.7RR162 pKa = 11.84AVRR165 pKa = 11.84RR166 pKa = 11.84QLRR169 pKa = 11.84TGVQPIQPVPHH180 pKa = 6.27VEE182 pKa = 4.12STHH185 pKa = 6.73RR186 pKa = 11.84SSDD189 pKa = 3.56PQLLEE194 pKa = 4.23SSTTCSANLQNDD206 pKa = 3.98GAGRR210 pKa = 11.84VIGGGIPTAIPEE222 pKa = 4.52GSDD225 pKa = 3.41VEE227 pKa = 4.66QVCHH231 pKa = 6.42ASHH234 pKa = 6.62YY235 pKa = 10.39GG236 pKa = 3.23
MM1 pKa = 7.66AVGKK5 pKa = 10.66YY6 pKa = 6.42MTIIINVNNDD16 pKa = 2.65EE17 pKa = 4.23RR18 pKa = 11.84KK19 pKa = 9.36QPEE22 pKa = 4.32GATGSSVRR30 pKa = 11.84RR31 pKa = 11.84GDD33 pKa = 3.53NKK35 pKa = 9.44RR36 pKa = 11.84TRR38 pKa = 11.84GNKK41 pKa = 8.59PRR43 pKa = 11.84SHH45 pKa = 6.71HH46 pKa = 5.96PGSRR50 pKa = 11.84EE51 pKa = 3.51RR52 pKa = 11.84KK53 pKa = 8.9GYY55 pKa = 9.09NHH57 pKa = 7.64PSPTPKK63 pKa = 10.09NSKK66 pKa = 9.6QGQLRR71 pKa = 11.84TEE73 pKa = 4.48AVQEE77 pKa = 4.25HH78 pKa = 7.11PKK80 pKa = 10.66HH81 pKa = 6.2GGTAFRR87 pKa = 11.84RR88 pKa = 11.84EE89 pKa = 4.5SGGSVHH95 pKa = 7.01PSHH98 pKa = 7.17PRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84ARR104 pKa = 11.84RR105 pKa = 11.84GGDD108 pKa = 3.06MAPRR112 pKa = 11.84QHH114 pKa = 6.96PPPPRR119 pKa = 11.84EE120 pKa = 3.54RR121 pKa = 11.84RR122 pKa = 11.84TKK124 pKa = 10.5TEE126 pKa = 3.69TQAEE130 pKa = 4.04RR131 pKa = 11.84RR132 pKa = 11.84AQALSVLPTLLDD144 pKa = 4.38CIGGLDD150 pKa = 3.94LGPAEE155 pKa = 4.56VLLHH159 pKa = 5.4CHH161 pKa = 5.7RR162 pKa = 11.84AVRR165 pKa = 11.84RR166 pKa = 11.84QLRR169 pKa = 11.84TGVQPIQPVPHH180 pKa = 6.27VEE182 pKa = 4.12STHH185 pKa = 6.73RR186 pKa = 11.84SSDD189 pKa = 3.56PQLLEE194 pKa = 4.23SSTTCSANLQNDD206 pKa = 3.98GAGRR210 pKa = 11.84VIGGGIPTAIPEE222 pKa = 4.52GSDD225 pKa = 3.41VEE227 pKa = 4.66QVCHH231 pKa = 6.42ASHH234 pKa = 6.62YY235 pKa = 10.39GG236 pKa = 3.23
Molecular weight: 25.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369 |
236 |
581 |
342.3 |
37.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.889 ± 1.406 | 2.63 ± 0.486 |
4.383 ± 0.626 | 5.771 ± 1.039 |
3.214 ± 1.031 | 6.866 ± 1.024 |
2.557 ± 0.839 | 3.871 ± 0.257 |
4.748 ± 0.731 | 8.327 ± 0.958 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.293 | 3.579 ± 0.617 |
8.035 ± 0.473 | 3.287 ± 0.559 |
7.962 ± 1.103 | 6.939 ± 0.823 |
5.844 ± 0.714 | 7.962 ± 0.618 |
1.461 ± 0.447 | 2.337 ± 0.531 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
