
Wenzhou narna-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.63
Get precalculated fractions of proteins
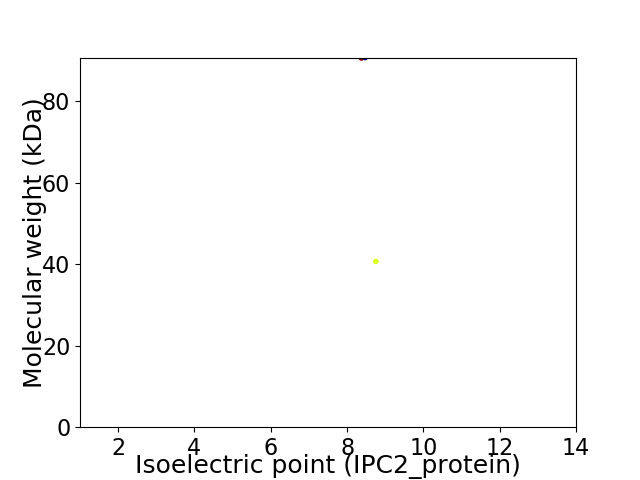
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KIS9|A0A1L3KIS9_9VIRU Capsid protein OS=Wenzhou narna-like virus 5 OX=1923580 PE=3 SV=1
MM1 pKa = 7.52SPLEE5 pKa = 4.26EE6 pKa = 4.47KK7 pKa = 10.68SSQKK11 pKa = 10.24RR12 pKa = 11.84IRR14 pKa = 11.84IKK16 pKa = 7.7THH18 pKa = 5.89EE19 pKa = 4.26FTSCGSTRR27 pKa = 11.84SFDD30 pKa = 3.4SGKK33 pKa = 10.19SLRR36 pKa = 11.84AFGSKK41 pKa = 9.79GAASMSRR48 pKa = 11.84AKK50 pKa = 10.76CRR52 pKa = 11.84DD53 pKa = 3.31SEE55 pKa = 4.14KK56 pKa = 11.09SKK58 pKa = 10.78ISVHH62 pKa = 4.16TTILSVLNKK71 pKa = 9.69VQTNNTEE78 pKa = 3.93VNVILEE84 pKa = 4.16SLLWPFVARR93 pKa = 11.84RR94 pKa = 11.84SQRR97 pKa = 11.84SEE99 pKa = 3.57MFSPKK104 pKa = 10.19EE105 pKa = 3.7FDD107 pKa = 3.5RR108 pKa = 11.84MVSSFKK114 pKa = 10.16KK115 pKa = 9.0TGEE118 pKa = 4.2TILKK122 pKa = 9.71YY123 pKa = 10.8VSADD127 pKa = 3.0NKK129 pKa = 8.15EE130 pKa = 4.13QKK132 pKa = 9.22FCKK135 pKa = 9.88YY136 pKa = 8.97WLDD139 pKa = 3.64YY140 pKa = 10.82YY141 pKa = 10.55MCLVFQDD148 pKa = 3.59HH149 pKa = 5.79QRR151 pKa = 11.84PEE153 pKa = 3.93KK154 pKa = 10.65PEE156 pKa = 3.89WLNSNLFSGWLKK168 pKa = 10.76RR169 pKa = 11.84FVCRR173 pKa = 11.84AIAKK177 pKa = 9.97DD178 pKa = 3.81DD179 pKa = 3.4VSFIYY184 pKa = 10.5SLQKK188 pKa = 9.97GSKK191 pKa = 9.4RR192 pKa = 11.84LWPRR196 pKa = 11.84LSKK199 pKa = 10.52QSEE202 pKa = 3.71YY203 pKa = 11.16DD204 pKa = 3.05AYY206 pKa = 10.88LKK208 pKa = 10.56HH209 pKa = 6.79ASRR212 pKa = 11.84LSSFHH217 pKa = 6.72GFLPDD222 pKa = 5.12DD223 pKa = 4.37LDD225 pKa = 4.11EE226 pKa = 5.16EE227 pKa = 4.54ISRR230 pKa = 11.84TSQEE234 pKa = 3.1IFLPINSGKK243 pKa = 9.19MEE245 pKa = 4.33ASKK248 pKa = 11.03YY249 pKa = 10.0CPTGSGCLQATKK261 pKa = 11.05NEE263 pKa = 3.97FGALSLLPTHH273 pKa = 6.84FEE275 pKa = 4.13VSDD278 pKa = 4.98DD279 pKa = 3.7IPNASSLGKK288 pKa = 10.18LRR290 pKa = 11.84ALNHH294 pKa = 5.88SFIKK298 pKa = 10.39YY299 pKa = 9.12RR300 pKa = 11.84QEE302 pKa = 4.82SYY304 pKa = 11.51DD305 pKa = 3.48QMLSKK310 pKa = 10.42VHH312 pKa = 6.53KK313 pKa = 9.88SLVDD317 pKa = 3.31AEE319 pKa = 5.1SIFDD323 pKa = 4.04LKK325 pKa = 10.74CVAVAEE331 pKa = 4.21PSKK334 pKa = 10.92FRR336 pKa = 11.84MITKK340 pKa = 10.3GDD342 pKa = 3.54GYY344 pKa = 10.75LYY346 pKa = 10.12TALQPLQGQMLDD358 pKa = 3.12CWKK361 pKa = 10.1KK362 pKa = 10.16SKK364 pKa = 10.74YY365 pKa = 9.31STMLEE370 pKa = 3.59QDD372 pKa = 3.27FEE374 pKa = 4.52KK375 pKa = 10.6RR376 pKa = 11.84IQTIDD381 pKa = 3.28KK382 pKa = 9.36NCRR385 pKa = 11.84ILEE388 pKa = 4.22FWCSVDD394 pKa = 4.11YY395 pKa = 10.8EE396 pKa = 4.32AATDD400 pKa = 4.92LIKK403 pKa = 10.9KK404 pKa = 8.89QATLTAIKK412 pKa = 10.14EE413 pKa = 3.93IDD415 pKa = 3.34HH416 pKa = 6.79KK417 pKa = 11.11YY418 pKa = 10.96SEE420 pKa = 4.45LAFLALSASGQCQYY434 pKa = 10.76PQLKK438 pKa = 9.55GFPKK442 pKa = 10.09IEE444 pKa = 5.8DD445 pKa = 3.39ITCIDD450 pKa = 4.06SQLMGHH456 pKa = 6.95PLSFPLLCTINLAVFRR472 pKa = 11.84CALKK476 pKa = 10.42RR477 pKa = 11.84WVNDD481 pKa = 3.32SPNSFEE487 pKa = 3.88KK488 pKa = 10.36RR489 pKa = 11.84CRR491 pKa = 11.84KK492 pKa = 8.9EE493 pKa = 3.68RR494 pKa = 11.84AEE496 pKa = 3.94LMYY499 pKa = 11.35NNVLVNGDD507 pKa = 3.99DD508 pKa = 3.69MLFKK512 pKa = 10.75CEE514 pKa = 3.63RR515 pKa = 11.84SFYY518 pKa = 10.52EE519 pKa = 3.97IFVKK523 pKa = 9.89TALDD527 pKa = 3.89CGLKK531 pKa = 9.55MSQGKK536 pKa = 9.68QYY538 pKa = 11.16LSVDD542 pKa = 2.85HH543 pKa = 6.92CMINSLVFKK552 pKa = 10.69RR553 pKa = 11.84VGNLMRR559 pKa = 11.84NQGYY563 pKa = 10.53LNLKK567 pKa = 9.35VISNEE572 pKa = 3.89SLKK575 pKa = 11.21DD576 pKa = 3.79GVSLCLPTQISKK588 pKa = 9.16TINEE592 pKa = 4.31MCSLVPWACCIIPTVFKK609 pKa = 10.51RR610 pKa = 11.84WSSDD614 pKa = 3.11LKK616 pKa = 10.98FLNFSPNWFLPCHH629 pKa = 6.46LGGFGVEE636 pKa = 3.75RR637 pKa = 11.84RR638 pKa = 11.84FGSVFKK644 pKa = 9.85LTRR647 pKa = 11.84QQRR650 pKa = 11.84KK651 pKa = 6.94VCTMFVKK658 pKa = 10.62NPEE661 pKa = 3.73LSLYY665 pKa = 6.99FTKK668 pKa = 10.7GFSLDD673 pKa = 3.06HH674 pKa = 6.44FYY676 pKa = 11.29KK677 pKa = 10.72FKK679 pKa = 10.96SQISGALSHH688 pKa = 6.95PRR690 pKa = 11.84FKK692 pKa = 10.73QSYY695 pKa = 9.7CPLNDD700 pKa = 4.02NEE702 pKa = 5.01CSDD705 pKa = 4.81DD706 pKa = 3.43PWLEE710 pKa = 4.26RR711 pKa = 11.84ISLIHH716 pKa = 6.54RR717 pKa = 11.84FSSFALRR724 pKa = 11.84KK725 pKa = 10.21NEE727 pKa = 3.91VAKK730 pKa = 10.8LRR732 pKa = 11.84EE733 pKa = 4.16FGKK736 pKa = 9.87EE737 pKa = 3.56LYY739 pKa = 10.14RR740 pKa = 11.84FRR742 pKa = 11.84TMPRR746 pKa = 11.84EE747 pKa = 3.95KK748 pKa = 10.72VDD750 pKa = 3.52TLVHH754 pKa = 6.22LKK756 pKa = 10.08FVSIKK761 pKa = 10.91GPICPPLPPLKK772 pKa = 10.55LRR774 pKa = 11.84TLNSHH779 pKa = 6.45IEE781 pKa = 4.11SSEE784 pKa = 4.0FPP786 pKa = 3.72
MM1 pKa = 7.52SPLEE5 pKa = 4.26EE6 pKa = 4.47KK7 pKa = 10.68SSQKK11 pKa = 10.24RR12 pKa = 11.84IRR14 pKa = 11.84IKK16 pKa = 7.7THH18 pKa = 5.89EE19 pKa = 4.26FTSCGSTRR27 pKa = 11.84SFDD30 pKa = 3.4SGKK33 pKa = 10.19SLRR36 pKa = 11.84AFGSKK41 pKa = 9.79GAASMSRR48 pKa = 11.84AKK50 pKa = 10.76CRR52 pKa = 11.84DD53 pKa = 3.31SEE55 pKa = 4.14KK56 pKa = 11.09SKK58 pKa = 10.78ISVHH62 pKa = 4.16TTILSVLNKK71 pKa = 9.69VQTNNTEE78 pKa = 3.93VNVILEE84 pKa = 4.16SLLWPFVARR93 pKa = 11.84RR94 pKa = 11.84SQRR97 pKa = 11.84SEE99 pKa = 3.57MFSPKK104 pKa = 10.19EE105 pKa = 3.7FDD107 pKa = 3.5RR108 pKa = 11.84MVSSFKK114 pKa = 10.16KK115 pKa = 9.0TGEE118 pKa = 4.2TILKK122 pKa = 9.71YY123 pKa = 10.8VSADD127 pKa = 3.0NKK129 pKa = 8.15EE130 pKa = 4.13QKK132 pKa = 9.22FCKK135 pKa = 9.88YY136 pKa = 8.97WLDD139 pKa = 3.64YY140 pKa = 10.82YY141 pKa = 10.55MCLVFQDD148 pKa = 3.59HH149 pKa = 5.79QRR151 pKa = 11.84PEE153 pKa = 3.93KK154 pKa = 10.65PEE156 pKa = 3.89WLNSNLFSGWLKK168 pKa = 10.76RR169 pKa = 11.84FVCRR173 pKa = 11.84AIAKK177 pKa = 9.97DD178 pKa = 3.81DD179 pKa = 3.4VSFIYY184 pKa = 10.5SLQKK188 pKa = 9.97GSKK191 pKa = 9.4RR192 pKa = 11.84LWPRR196 pKa = 11.84LSKK199 pKa = 10.52QSEE202 pKa = 3.71YY203 pKa = 11.16DD204 pKa = 3.05AYY206 pKa = 10.88LKK208 pKa = 10.56HH209 pKa = 6.79ASRR212 pKa = 11.84LSSFHH217 pKa = 6.72GFLPDD222 pKa = 5.12DD223 pKa = 4.37LDD225 pKa = 4.11EE226 pKa = 5.16EE227 pKa = 4.54ISRR230 pKa = 11.84TSQEE234 pKa = 3.1IFLPINSGKK243 pKa = 9.19MEE245 pKa = 4.33ASKK248 pKa = 11.03YY249 pKa = 10.0CPTGSGCLQATKK261 pKa = 11.05NEE263 pKa = 3.97FGALSLLPTHH273 pKa = 6.84FEE275 pKa = 4.13VSDD278 pKa = 4.98DD279 pKa = 3.7IPNASSLGKK288 pKa = 10.18LRR290 pKa = 11.84ALNHH294 pKa = 5.88SFIKK298 pKa = 10.39YY299 pKa = 9.12RR300 pKa = 11.84QEE302 pKa = 4.82SYY304 pKa = 11.51DD305 pKa = 3.48QMLSKK310 pKa = 10.42VHH312 pKa = 6.53KK313 pKa = 9.88SLVDD317 pKa = 3.31AEE319 pKa = 5.1SIFDD323 pKa = 4.04LKK325 pKa = 10.74CVAVAEE331 pKa = 4.21PSKK334 pKa = 10.92FRR336 pKa = 11.84MITKK340 pKa = 10.3GDD342 pKa = 3.54GYY344 pKa = 10.75LYY346 pKa = 10.12TALQPLQGQMLDD358 pKa = 3.12CWKK361 pKa = 10.1KK362 pKa = 10.16SKK364 pKa = 10.74YY365 pKa = 9.31STMLEE370 pKa = 3.59QDD372 pKa = 3.27FEE374 pKa = 4.52KK375 pKa = 10.6RR376 pKa = 11.84IQTIDD381 pKa = 3.28KK382 pKa = 9.36NCRR385 pKa = 11.84ILEE388 pKa = 4.22FWCSVDD394 pKa = 4.11YY395 pKa = 10.8EE396 pKa = 4.32AATDD400 pKa = 4.92LIKK403 pKa = 10.9KK404 pKa = 8.89QATLTAIKK412 pKa = 10.14EE413 pKa = 3.93IDD415 pKa = 3.34HH416 pKa = 6.79KK417 pKa = 11.11YY418 pKa = 10.96SEE420 pKa = 4.45LAFLALSASGQCQYY434 pKa = 10.76PQLKK438 pKa = 9.55GFPKK442 pKa = 10.09IEE444 pKa = 5.8DD445 pKa = 3.39ITCIDD450 pKa = 4.06SQLMGHH456 pKa = 6.95PLSFPLLCTINLAVFRR472 pKa = 11.84CALKK476 pKa = 10.42RR477 pKa = 11.84WVNDD481 pKa = 3.32SPNSFEE487 pKa = 3.88KK488 pKa = 10.36RR489 pKa = 11.84CRR491 pKa = 11.84KK492 pKa = 8.9EE493 pKa = 3.68RR494 pKa = 11.84AEE496 pKa = 3.94LMYY499 pKa = 11.35NNVLVNGDD507 pKa = 3.99DD508 pKa = 3.69MLFKK512 pKa = 10.75CEE514 pKa = 3.63RR515 pKa = 11.84SFYY518 pKa = 10.52EE519 pKa = 3.97IFVKK523 pKa = 9.89TALDD527 pKa = 3.89CGLKK531 pKa = 9.55MSQGKK536 pKa = 9.68QYY538 pKa = 11.16LSVDD542 pKa = 2.85HH543 pKa = 6.92CMINSLVFKK552 pKa = 10.69RR553 pKa = 11.84VGNLMRR559 pKa = 11.84NQGYY563 pKa = 10.53LNLKK567 pKa = 9.35VISNEE572 pKa = 3.89SLKK575 pKa = 11.21DD576 pKa = 3.79GVSLCLPTQISKK588 pKa = 9.16TINEE592 pKa = 4.31MCSLVPWACCIIPTVFKK609 pKa = 10.51RR610 pKa = 11.84WSSDD614 pKa = 3.11LKK616 pKa = 10.98FLNFSPNWFLPCHH629 pKa = 6.46LGGFGVEE636 pKa = 3.75RR637 pKa = 11.84RR638 pKa = 11.84FGSVFKK644 pKa = 9.85LTRR647 pKa = 11.84QQRR650 pKa = 11.84KK651 pKa = 6.94VCTMFVKK658 pKa = 10.62NPEE661 pKa = 3.73LSLYY665 pKa = 6.99FTKK668 pKa = 10.7GFSLDD673 pKa = 3.06HH674 pKa = 6.44FYY676 pKa = 11.29KK677 pKa = 10.72FKK679 pKa = 10.96SQISGALSHH688 pKa = 6.95PRR690 pKa = 11.84FKK692 pKa = 10.73QSYY695 pKa = 9.7CPLNDD700 pKa = 4.02NEE702 pKa = 5.01CSDD705 pKa = 4.81DD706 pKa = 3.43PWLEE710 pKa = 4.26RR711 pKa = 11.84ISLIHH716 pKa = 6.54RR717 pKa = 11.84FSSFALRR724 pKa = 11.84KK725 pKa = 10.21NEE727 pKa = 3.91VAKK730 pKa = 10.8LRR732 pKa = 11.84EE733 pKa = 4.16FGKK736 pKa = 9.87EE737 pKa = 3.56LYY739 pKa = 10.14RR740 pKa = 11.84FRR742 pKa = 11.84TMPRR746 pKa = 11.84EE747 pKa = 3.95KK748 pKa = 10.72VDD750 pKa = 3.52TLVHH754 pKa = 6.22LKK756 pKa = 10.08FVSIKK761 pKa = 10.91GPICPPLPPLKK772 pKa = 10.55LRR774 pKa = 11.84TLNSHH779 pKa = 6.45IEE781 pKa = 4.11SSEE784 pKa = 4.0FPP786 pKa = 3.72
Molecular weight: 90.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIS9|A0A1L3KIS9_9VIRU Capsid protein OS=Wenzhou narna-like virus 5 OX=1923580 PE=3 SV=1
MM1 pKa = 7.35YY2 pKa = 10.47SPSQSLKK9 pKa = 10.86HH10 pKa = 6.48PILMIKK16 pKa = 9.8SPKK19 pKa = 7.81TANVPMKK26 pKa = 9.41EE27 pKa = 3.87AKK29 pKa = 9.2PRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84SRR36 pKa = 11.84PKK38 pKa = 10.01QPQQKK43 pKa = 9.5LQTEE47 pKa = 4.48INEE50 pKa = 4.2MAPVSYY56 pKa = 10.95ASTRR60 pKa = 11.84SFRR63 pKa = 11.84TDD65 pKa = 2.59VLQSHH70 pKa = 6.91KK71 pKa = 10.35NVKK74 pKa = 9.92SRR76 pKa = 11.84EE77 pKa = 4.09QISSITGSNSFLVGGTFHH95 pKa = 7.43INPGLPEE102 pKa = 4.41CFPYY106 pKa = 10.93LSDD109 pKa = 3.41IAKK112 pKa = 10.19KK113 pKa = 8.75FQQYY117 pKa = 9.22RR118 pKa = 11.84FKK120 pKa = 11.1SLTFEE125 pKa = 4.65FVTSTATTTTGNVILSPEE143 pKa = 4.26YY144 pKa = 10.31NPKK147 pKa = 9.89EE148 pKa = 3.96LPPATEE154 pKa = 3.84QQATNTYY161 pKa = 8.96GAVEE165 pKa = 4.3SVCWRR170 pKa = 11.84NSKK173 pKa = 10.31VSFSPSGMFPLGPRR187 pKa = 11.84KK188 pKa = 9.23MIRR191 pKa = 11.84SCLVPSDD198 pKa = 3.88LQAYY202 pKa = 8.53DD203 pKa = 3.29AGQLFVCVTGQTGNPLIGKK222 pKa = 8.63LWVSYY227 pKa = 10.92DD228 pKa = 3.0VDD230 pKa = 4.86LYY232 pKa = 10.73IPQNSLVQDD241 pKa = 3.82YY242 pKa = 9.99SPSSAAMFTIVSTALTSGVATLLTLTTVYY271 pKa = 11.12DD272 pKa = 3.77PFGFTNNGNTLSLLKK287 pKa = 10.4GVYY290 pKa = 10.3RR291 pKa = 11.84MDD293 pKa = 3.45ARR295 pKa = 11.84FTVDD299 pKa = 3.1GASVTNCLTQFSVGGVLSTTHH320 pKa = 6.57TYY322 pKa = 10.33VAGSPTLWTYY332 pKa = 10.95FGTGLQMLIPSTEE345 pKa = 3.88VQNFGISVASTFTGAANAGNGVLIINLVV373 pKa = 3.2
MM1 pKa = 7.35YY2 pKa = 10.47SPSQSLKK9 pKa = 10.86HH10 pKa = 6.48PILMIKK16 pKa = 9.8SPKK19 pKa = 7.81TANVPMKK26 pKa = 9.41EE27 pKa = 3.87AKK29 pKa = 9.2PRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84SRR36 pKa = 11.84PKK38 pKa = 10.01QPQQKK43 pKa = 9.5LQTEE47 pKa = 4.48INEE50 pKa = 4.2MAPVSYY56 pKa = 10.95ASTRR60 pKa = 11.84SFRR63 pKa = 11.84TDD65 pKa = 2.59VLQSHH70 pKa = 6.91KK71 pKa = 10.35NVKK74 pKa = 9.92SRR76 pKa = 11.84EE77 pKa = 4.09QISSITGSNSFLVGGTFHH95 pKa = 7.43INPGLPEE102 pKa = 4.41CFPYY106 pKa = 10.93LSDD109 pKa = 3.41IAKK112 pKa = 10.19KK113 pKa = 8.75FQQYY117 pKa = 9.22RR118 pKa = 11.84FKK120 pKa = 11.1SLTFEE125 pKa = 4.65FVTSTATTTTGNVILSPEE143 pKa = 4.26YY144 pKa = 10.31NPKK147 pKa = 9.89EE148 pKa = 3.96LPPATEE154 pKa = 3.84QQATNTYY161 pKa = 8.96GAVEE165 pKa = 4.3SVCWRR170 pKa = 11.84NSKK173 pKa = 10.31VSFSPSGMFPLGPRR187 pKa = 11.84KK188 pKa = 9.23MIRR191 pKa = 11.84SCLVPSDD198 pKa = 3.88LQAYY202 pKa = 8.53DD203 pKa = 3.29AGQLFVCVTGQTGNPLIGKK222 pKa = 8.63LWVSYY227 pKa = 10.92DD228 pKa = 3.0VDD230 pKa = 4.86LYY232 pKa = 10.73IPQNSLVQDD241 pKa = 3.82YY242 pKa = 9.99SPSSAAMFTIVSTALTSGVATLLTLTTVYY271 pKa = 11.12DD272 pKa = 3.77PFGFTNNGNTLSLLKK287 pKa = 10.4GVYY290 pKa = 10.3RR291 pKa = 11.84MDD293 pKa = 3.45ARR295 pKa = 11.84FTVDD299 pKa = 3.1GASVTNCLTQFSVGGVLSTTHH320 pKa = 6.57TYY322 pKa = 10.33VAGSPTLWTYY332 pKa = 10.95FGTGLQMLIPSTEE345 pKa = 3.88VQNFGISVASTFTGAANAGNGVLIINLVV373 pKa = 3.2
Molecular weight: 40.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1159 |
373 |
786 |
579.5 |
65.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.832 ± 0.594 | 2.847 ± 0.839 |
4.142 ± 0.814 | 4.918 ± 1.097 |
5.953 ± 0.479 | 5.091 ± 1.047 |
1.812 ± 0.412 | 4.659 ± 0.206 |
7.593 ± 1.691 | 10.009 ± 0.796 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.416 ± 0.002 | 4.314 ± 0.434 |
5.091 ± 0.898 | 4.228 ± 0.482 |
5.004 ± 0.697 | 10.699 ± 0.014 |
6.126 ± 2.412 | 5.695 ± 1.159 |
1.294 ± 0.273 | 3.279 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
