
Alteromonadaceae bacterium 2052S.S.stab0a.01
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; unclassified Alteromonadaceae
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
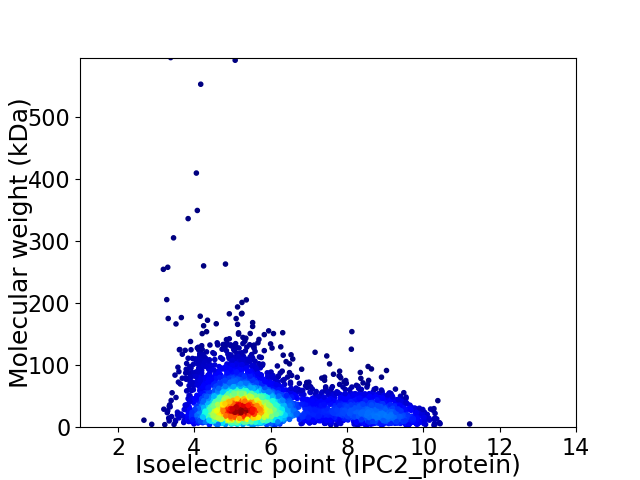
Virtual 2D-PAGE plot for 4578 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495D8X9|A0A495D8X9_9ALTE Uncharacterized protein OS=Alteromonadaceae bacterium 2052S.S.stab0a.01 OX=1304900 GN=K256_1559 PE=4 SV=1
MM1 pKa = 7.31IQQNYY6 pKa = 8.48LFRR9 pKa = 11.84RR10 pKa = 11.84AAPVSALLLGSLFSLASTQALALQCSYY37 pKa = 10.87EE38 pKa = 3.99VSNDD42 pKa = 2.88WGSGFTANITINNDD56 pKa = 2.48GTTTVSDD63 pKa = 4.7WEE65 pKa = 4.55VQWQQNGGSTVTGLWNAAYY84 pKa = 8.73TGSNPYY90 pKa = 8.92TATDD94 pKa = 3.69AGWNANIAPGSSASFGFQGNGEE116 pKa = 4.52DD117 pKa = 3.49STGTILSCTAGGSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSGGNSTTLVIQEE208 pKa = 4.12DD209 pKa = 3.93TGLCTVDD216 pKa = 5.16GSIDD220 pKa = 3.72SNNAGFTGSGFANTDD235 pKa = 3.02NVIGSRR241 pKa = 11.84VEE243 pKa = 3.79YY244 pKa = 10.43SIYY247 pKa = 10.64AAAAGNVLLDD257 pKa = 3.43ARR259 pKa = 11.84YY260 pKa = 7.82ATTSSRR266 pKa = 11.84AGDD269 pKa = 3.59VEE271 pKa = 4.37INGVNVGAFDD281 pKa = 4.55FVSTGDD287 pKa = 3.22WTSWTTEE294 pKa = 4.2SINVPLQAGLNSVSLIATTDD314 pKa = 3.13SGLPNIDD321 pKa = 3.63SLTAIGNDD329 pKa = 4.31LSAGDD334 pKa = 4.63CGSGSSSSSSSSSSSGSSSSGGVFNTLVIEE364 pKa = 4.15EE365 pKa = 4.72EE366 pKa = 3.87EE367 pKa = 3.98AGYY370 pKa = 10.97CNTLSVPVEE379 pKa = 4.04NEE381 pKa = 3.67HH382 pKa = 6.59TGFTGSGYY390 pKa = 11.1ANSEE394 pKa = 4.09NATGTGLEE402 pKa = 4.23WQVTTGEE409 pKa = 4.57AGTATIDD416 pKa = 3.14VRR418 pKa = 11.84FANGSDD424 pKa = 3.32ADD426 pKa = 4.22RR427 pKa = 11.84PGTLYY432 pKa = 10.57INGGADD438 pKa = 3.24GAYY441 pKa = 9.17TISLPATGAWTTWQTSSISVPLPAGDD467 pKa = 4.2SSLEE471 pKa = 3.97LLADD475 pKa = 3.97TANGLANIDD484 pKa = 4.06SLTITALDD492 pKa = 4.08VSPLDD497 pKa = 3.98CAEE500 pKa = 4.45VPPEE504 pKa = 4.24VPDD507 pKa = 3.64EE508 pKa = 4.16LLAFPGAQGFGRR520 pKa = 11.84LAVGGRR526 pKa = 11.84YY527 pKa = 10.06GEE529 pKa = 4.46VVHH532 pKa = 5.63VTNLNDD538 pKa = 3.5SGTGSLRR545 pKa = 11.84DD546 pKa = 4.83AISQPNRR553 pKa = 11.84IVVFDD558 pKa = 3.53VGGVINLSSRR568 pKa = 11.84LVFKK572 pKa = 11.04NNQTIAGQTAPGGGITLYY590 pKa = 11.34GNGTSFSAASNTIVRR605 pKa = 11.84YY606 pKa = 9.42VRR608 pKa = 11.84FRR610 pKa = 11.84MGINGDD616 pKa = 3.75SGKK619 pKa = 9.05DD620 pKa = 3.49TITMANGHH628 pKa = 5.61DD629 pKa = 4.57VIWDD633 pKa = 3.6HH634 pKa = 7.4CSLSWGRR641 pKa = 11.84DD642 pKa = 2.94GTLDD646 pKa = 3.73LNQEE650 pKa = 4.39SGSTLYY656 pKa = 11.22NLTLQDD662 pKa = 3.93SIVAQGLQTHH672 pKa = 5.35STGGLVNTTGTSIIRR687 pKa = 11.84SLYY690 pKa = 9.43IDD692 pKa = 3.54NNSRR696 pKa = 11.84NPKK699 pKa = 10.16ARR701 pKa = 11.84GTLQFVNNVLYY712 pKa = 10.7NWVTSGYY719 pKa = 10.21ILGDD723 pKa = 3.17TSGRR727 pKa = 11.84SDD729 pKa = 2.98GAMIGNFLIAGPEE742 pKa = 4.32TNGGTLSSPTPEE754 pKa = 3.88YY755 pKa = 10.9NIYY758 pKa = 10.81AVDD761 pKa = 2.96NWYY764 pKa = 10.57DD765 pKa = 3.6NNKK768 pKa = 10.68DD769 pKa = 3.05GTLNARR775 pKa = 11.84VLGEE779 pKa = 4.18GDD781 pKa = 3.49FGSATWWTTPSVEE794 pKa = 4.36YY795 pKa = 8.75PTVPALSADD804 pKa = 3.27AALQYY809 pKa = 10.67VINNAGTSQWRR820 pKa = 11.84DD821 pKa = 3.17EE822 pKa = 4.2VDD824 pKa = 3.06QYY826 pKa = 12.13LMDD829 pKa = 5.0EE830 pKa = 4.2LTSWGRR836 pKa = 11.84QGATISNEE844 pKa = 3.82SSLGIPNNVGVIPGGTAPTDD864 pKa = 3.41TDD866 pKa = 3.86RR867 pKa = 11.84DD868 pKa = 3.77GMPDD872 pKa = 2.85SWEE875 pKa = 4.0TQMGFNPNNAADD887 pKa = 4.03AFADD891 pKa = 3.62ADD893 pKa = 3.73GDD895 pKa = 3.68GWVNLEE901 pKa = 4.18EE902 pKa = 4.66YY903 pKa = 10.42INSLVPP909 pKa = 3.73
MM1 pKa = 7.31IQQNYY6 pKa = 8.48LFRR9 pKa = 11.84RR10 pKa = 11.84AAPVSALLLGSLFSLASTQALALQCSYY37 pKa = 10.87EE38 pKa = 3.99VSNDD42 pKa = 2.88WGSGFTANITINNDD56 pKa = 2.48GTTTVSDD63 pKa = 4.7WEE65 pKa = 4.55VQWQQNGGSTVTGLWNAAYY84 pKa = 8.73TGSNPYY90 pKa = 8.92TATDD94 pKa = 3.69AGWNANIAPGSSASFGFQGNGEE116 pKa = 4.52DD117 pKa = 3.49STGTILSCTAGGSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSGGNSTTLVIQEE208 pKa = 4.12DD209 pKa = 3.93TGLCTVDD216 pKa = 5.16GSIDD220 pKa = 3.72SNNAGFTGSGFANTDD235 pKa = 3.02NVIGSRR241 pKa = 11.84VEE243 pKa = 3.79YY244 pKa = 10.43SIYY247 pKa = 10.64AAAAGNVLLDD257 pKa = 3.43ARR259 pKa = 11.84YY260 pKa = 7.82ATTSSRR266 pKa = 11.84AGDD269 pKa = 3.59VEE271 pKa = 4.37INGVNVGAFDD281 pKa = 4.55FVSTGDD287 pKa = 3.22WTSWTTEE294 pKa = 4.2SINVPLQAGLNSVSLIATTDD314 pKa = 3.13SGLPNIDD321 pKa = 3.63SLTAIGNDD329 pKa = 4.31LSAGDD334 pKa = 4.63CGSGSSSSSSSSSSSGSSSSGGVFNTLVIEE364 pKa = 4.15EE365 pKa = 4.72EE366 pKa = 3.87EE367 pKa = 3.98AGYY370 pKa = 10.97CNTLSVPVEE379 pKa = 4.04NEE381 pKa = 3.67HH382 pKa = 6.59TGFTGSGYY390 pKa = 11.1ANSEE394 pKa = 4.09NATGTGLEE402 pKa = 4.23WQVTTGEE409 pKa = 4.57AGTATIDD416 pKa = 3.14VRR418 pKa = 11.84FANGSDD424 pKa = 3.32ADD426 pKa = 4.22RR427 pKa = 11.84PGTLYY432 pKa = 10.57INGGADD438 pKa = 3.24GAYY441 pKa = 9.17TISLPATGAWTTWQTSSISVPLPAGDD467 pKa = 4.2SSLEE471 pKa = 3.97LLADD475 pKa = 3.97TANGLANIDD484 pKa = 4.06SLTITALDD492 pKa = 4.08VSPLDD497 pKa = 3.98CAEE500 pKa = 4.45VPPEE504 pKa = 4.24VPDD507 pKa = 3.64EE508 pKa = 4.16LLAFPGAQGFGRR520 pKa = 11.84LAVGGRR526 pKa = 11.84YY527 pKa = 10.06GEE529 pKa = 4.46VVHH532 pKa = 5.63VTNLNDD538 pKa = 3.5SGTGSLRR545 pKa = 11.84DD546 pKa = 4.83AISQPNRR553 pKa = 11.84IVVFDD558 pKa = 3.53VGGVINLSSRR568 pKa = 11.84LVFKK572 pKa = 11.04NNQTIAGQTAPGGGITLYY590 pKa = 11.34GNGTSFSAASNTIVRR605 pKa = 11.84YY606 pKa = 9.42VRR608 pKa = 11.84FRR610 pKa = 11.84MGINGDD616 pKa = 3.75SGKK619 pKa = 9.05DD620 pKa = 3.49TITMANGHH628 pKa = 5.61DD629 pKa = 4.57VIWDD633 pKa = 3.6HH634 pKa = 7.4CSLSWGRR641 pKa = 11.84DD642 pKa = 2.94GTLDD646 pKa = 3.73LNQEE650 pKa = 4.39SGSTLYY656 pKa = 11.22NLTLQDD662 pKa = 3.93SIVAQGLQTHH672 pKa = 5.35STGGLVNTTGTSIIRR687 pKa = 11.84SLYY690 pKa = 9.43IDD692 pKa = 3.54NNSRR696 pKa = 11.84NPKK699 pKa = 10.16ARR701 pKa = 11.84GTLQFVNNVLYY712 pKa = 10.7NWVTSGYY719 pKa = 10.21ILGDD723 pKa = 3.17TSGRR727 pKa = 11.84SDD729 pKa = 2.98GAMIGNFLIAGPEE742 pKa = 4.32TNGGTLSSPTPEE754 pKa = 3.88YY755 pKa = 10.9NIYY758 pKa = 10.81AVDD761 pKa = 2.96NWYY764 pKa = 10.57DD765 pKa = 3.6NNKK768 pKa = 10.68DD769 pKa = 3.05GTLNARR775 pKa = 11.84VLGEE779 pKa = 4.18GDD781 pKa = 3.49FGSATWWTTPSVEE794 pKa = 4.36YY795 pKa = 8.75PTVPALSADD804 pKa = 3.27AALQYY809 pKa = 10.67VINNAGTSQWRR820 pKa = 11.84DD821 pKa = 3.17EE822 pKa = 4.2VDD824 pKa = 3.06QYY826 pKa = 12.13LMDD829 pKa = 5.0EE830 pKa = 4.2LTSWGRR836 pKa = 11.84QGATISNEE844 pKa = 3.82SSLGIPNNVGVIPGGTAPTDD864 pKa = 3.41TDD866 pKa = 3.86RR867 pKa = 11.84DD868 pKa = 3.77GMPDD872 pKa = 2.85SWEE875 pKa = 4.0TQMGFNPNNAADD887 pKa = 4.03AFADD891 pKa = 3.62ADD893 pKa = 3.73GDD895 pKa = 3.68GWVNLEE901 pKa = 4.18EE902 pKa = 4.66YY903 pKa = 10.42INSLVPP909 pKa = 3.73
Molecular weight: 93.31 kDa
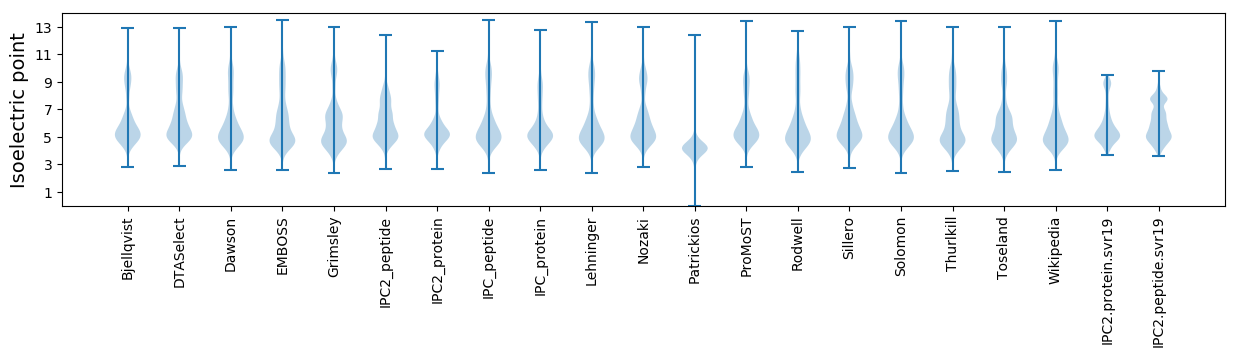
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A495DIU6|A0A495DIU6_9ALTE Cell migration-inducing and hyaluronan-binding protein OS=Alteromonadaceae bacterium 2052S.S.stab0a.01 OX=1304900 GN=K256_4544 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.31QLAAA44 pKa = 4.14
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.31QLAAA44 pKa = 4.14
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1627577 |
29 |
5920 |
355.5 |
39.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.466 ± 0.042 | 1.069 ± 0.014 |
5.919 ± 0.041 | 6.085 ± 0.036 |
3.95 ± 0.023 | 7.537 ± 0.04 |
2.186 ± 0.021 | 5.206 ± 0.026 |
3.792 ± 0.038 | 10.281 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.261 ± 0.017 | 3.908 ± 0.025 |
4.425 ± 0.031 | 4.258 ± 0.021 |
5.492 ± 0.033 | 7.047 ± 0.065 |
5.531 ± 0.036 | 7.091 ± 0.031 |
1.43 ± 0.015 | 3.065 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
