
Sewage-associated circular DNA virus-13
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
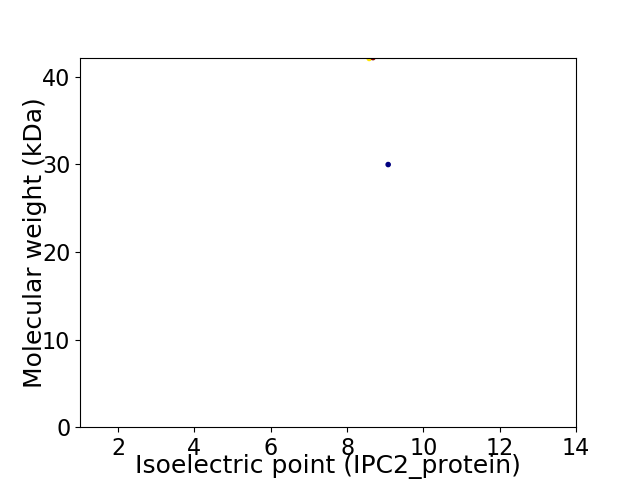
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075IZT3|A0A075IZT3_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-13 OX=1519389 PE=3 SV=1
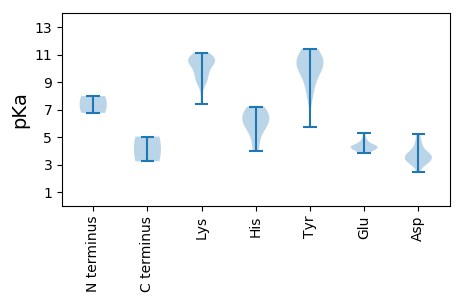
MM1 pKa = 7.98DD2 pKa = 5.23LPRR5 pKa = 11.84FRR7 pKa = 11.84LNNQRR12 pKa = 11.84LLCTYY17 pKa = 10.47AKK19 pKa = 10.52SDD21 pKa = 3.71LSKK24 pKa = 11.14KK25 pKa = 9.58KK26 pKa = 10.34LKK28 pKa = 10.54EE29 pKa = 3.83FFVSKK34 pKa = 10.55SPKK37 pKa = 8.77TSPKK41 pKa = 10.07VKK43 pKa = 9.79IARR46 pKa = 11.84EE47 pKa = 4.02SHH49 pKa = 6.34KK50 pKa = 11.0DD51 pKa = 3.39GSKK54 pKa = 10.47HH55 pKa = 4.0MHH57 pKa = 5.73VLVDD61 pKa = 3.88FGKK64 pKa = 10.8RR65 pKa = 11.84FDD67 pKa = 3.62STNRR71 pKa = 11.84RR72 pKa = 11.84VFDD75 pKa = 3.77FEE77 pKa = 4.27GRR79 pKa = 11.84HH80 pKa = 4.7PNIRR84 pKa = 11.84RR85 pKa = 11.84VEE87 pKa = 4.23SMIHH91 pKa = 5.58WANLLCYY98 pKa = 10.09LGKK101 pKa = 10.01EE102 pKa = 4.32DD103 pKa = 4.48KK104 pKa = 10.42KK105 pKa = 10.81CRR107 pKa = 11.84PSASVLAAAKK117 pKa = 9.5CWNAKK122 pKa = 8.92TPVEE126 pKa = 4.75AILSGAYY133 pKa = 9.31KK134 pKa = 10.67DD135 pKa = 4.61PIRR138 pKa = 11.84SLAVFRR144 pKa = 11.84QRR146 pKa = 11.84PSARR150 pKa = 11.84EE151 pKa = 3.92FICNMTEE158 pKa = 4.99PEE160 pKa = 4.24APWYY164 pKa = 9.84QDD166 pKa = 2.45IVKK169 pKa = 9.68PLIEE173 pKa = 4.86ANSDD177 pKa = 2.91HH178 pKa = 7.18RR179 pKa = 11.84SWYY182 pKa = 8.26WFCDD186 pKa = 3.16KK187 pKa = 11.03VGNSWKK193 pKa = 10.68SYY195 pKa = 5.69TCKK198 pKa = 10.71YY199 pKa = 10.21LLQTMRR205 pKa = 11.84EE206 pKa = 4.2DD207 pKa = 3.83FRR209 pKa = 11.84ATKK212 pKa = 9.14LTANVDD218 pKa = 3.68SFAMKK223 pKa = 10.31LGNWQQEE230 pKa = 4.09GWSGKK235 pKa = 9.8ILFVDD240 pKa = 4.54LARR243 pKa = 11.84GKK245 pKa = 10.98ANTNTIYY252 pKa = 10.78EE253 pKa = 4.42CLEE256 pKa = 4.02LTSDD260 pKa = 3.2GWGDD264 pKa = 3.25TGKK267 pKa = 10.79YY268 pKa = 8.49EE269 pKa = 4.35GKK271 pKa = 10.46SFILDD276 pKa = 3.44SSPIVVVLANWWPRR290 pKa = 11.84IAADD294 pKa = 3.62TGSLDD299 pKa = 2.95RR300 pKa = 11.84WKK302 pKa = 10.86LFEE305 pKa = 4.37ITNEE309 pKa = 4.29PIRR312 pKa = 11.84GSARR316 pKa = 11.84CMNLNIVRR324 pKa = 11.84TEE326 pKa = 3.85RR327 pKa = 11.84TDD329 pKa = 3.55LDD331 pKa = 3.83EE332 pKa = 4.61EE333 pKa = 4.23LAIQKK338 pKa = 9.39EE339 pKa = 4.42VARR342 pKa = 11.84LEE344 pKa = 4.33KK345 pKa = 10.46QMKK348 pKa = 9.27AAEE351 pKa = 4.13AVEE354 pKa = 4.09KK355 pKa = 10.89AKK357 pKa = 10.95ARR359 pKa = 11.84IAASRR364 pKa = 11.84DD365 pKa = 3.53CC366 pKa = 5.03
MM1 pKa = 7.98DD2 pKa = 5.23LPRR5 pKa = 11.84FRR7 pKa = 11.84LNNQRR12 pKa = 11.84LLCTYY17 pKa = 10.47AKK19 pKa = 10.52SDD21 pKa = 3.71LSKK24 pKa = 11.14KK25 pKa = 9.58KK26 pKa = 10.34LKK28 pKa = 10.54EE29 pKa = 3.83FFVSKK34 pKa = 10.55SPKK37 pKa = 8.77TSPKK41 pKa = 10.07VKK43 pKa = 9.79IARR46 pKa = 11.84EE47 pKa = 4.02SHH49 pKa = 6.34KK50 pKa = 11.0DD51 pKa = 3.39GSKK54 pKa = 10.47HH55 pKa = 4.0MHH57 pKa = 5.73VLVDD61 pKa = 3.88FGKK64 pKa = 10.8RR65 pKa = 11.84FDD67 pKa = 3.62STNRR71 pKa = 11.84RR72 pKa = 11.84VFDD75 pKa = 3.77FEE77 pKa = 4.27GRR79 pKa = 11.84HH80 pKa = 4.7PNIRR84 pKa = 11.84RR85 pKa = 11.84VEE87 pKa = 4.23SMIHH91 pKa = 5.58WANLLCYY98 pKa = 10.09LGKK101 pKa = 10.01EE102 pKa = 4.32DD103 pKa = 4.48KK104 pKa = 10.42KK105 pKa = 10.81CRR107 pKa = 11.84PSASVLAAAKK117 pKa = 9.5CWNAKK122 pKa = 8.92TPVEE126 pKa = 4.75AILSGAYY133 pKa = 9.31KK134 pKa = 10.67DD135 pKa = 4.61PIRR138 pKa = 11.84SLAVFRR144 pKa = 11.84QRR146 pKa = 11.84PSARR150 pKa = 11.84EE151 pKa = 3.92FICNMTEE158 pKa = 4.99PEE160 pKa = 4.24APWYY164 pKa = 9.84QDD166 pKa = 2.45IVKK169 pKa = 9.68PLIEE173 pKa = 4.86ANSDD177 pKa = 2.91HH178 pKa = 7.18RR179 pKa = 11.84SWYY182 pKa = 8.26WFCDD186 pKa = 3.16KK187 pKa = 11.03VGNSWKK193 pKa = 10.68SYY195 pKa = 5.69TCKK198 pKa = 10.71YY199 pKa = 10.21LLQTMRR205 pKa = 11.84EE206 pKa = 4.2DD207 pKa = 3.83FRR209 pKa = 11.84ATKK212 pKa = 9.14LTANVDD218 pKa = 3.68SFAMKK223 pKa = 10.31LGNWQQEE230 pKa = 4.09GWSGKK235 pKa = 9.8ILFVDD240 pKa = 4.54LARR243 pKa = 11.84GKK245 pKa = 10.98ANTNTIYY252 pKa = 10.78EE253 pKa = 4.42CLEE256 pKa = 4.02LTSDD260 pKa = 3.2GWGDD264 pKa = 3.25TGKK267 pKa = 10.79YY268 pKa = 8.49EE269 pKa = 4.35GKK271 pKa = 10.46SFILDD276 pKa = 3.44SSPIVVVLANWWPRR290 pKa = 11.84IAADD294 pKa = 3.62TGSLDD299 pKa = 2.95RR300 pKa = 11.84WKK302 pKa = 10.86LFEE305 pKa = 4.37ITNEE309 pKa = 4.29PIRR312 pKa = 11.84GSARR316 pKa = 11.84CMNLNIVRR324 pKa = 11.84TEE326 pKa = 3.85RR327 pKa = 11.84TDD329 pKa = 3.55LDD331 pKa = 3.83EE332 pKa = 4.61EE333 pKa = 4.23LAIQKK338 pKa = 9.39EE339 pKa = 4.42VARR342 pKa = 11.84LEE344 pKa = 4.33KK345 pKa = 10.46QMKK348 pKa = 9.27AAEE351 pKa = 4.13AVEE354 pKa = 4.09KK355 pKa = 10.89AKK357 pKa = 10.95ARR359 pKa = 11.84IAASRR364 pKa = 11.84DD365 pKa = 3.53CC366 pKa = 5.03
Molecular weight: 42.09 kDa
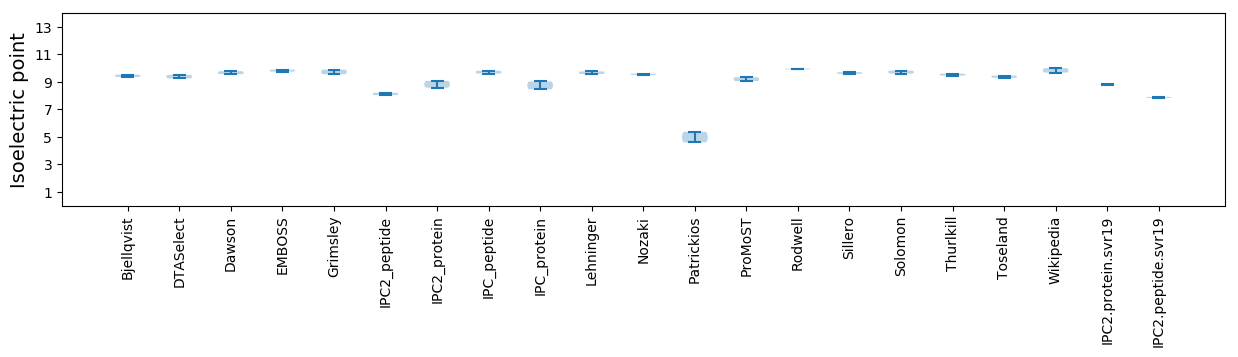
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075IZT3|A0A075IZT3_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-13 OX=1519389 PE=3 SV=1
MM1 pKa = 6.76STKK4 pKa = 10.24KK5 pKa = 9.31RR6 pKa = 11.84TAVPARR12 pKa = 11.84KK13 pKa = 7.38TVAAPKK19 pKa = 8.84RR20 pKa = 11.84TYY22 pKa = 11.1AKK24 pKa = 8.92RR25 pKa = 11.84TYY27 pKa = 7.88TARR30 pKa = 11.84APAATRR36 pKa = 11.84YY37 pKa = 9.79RR38 pKa = 11.84GTGDD42 pKa = 3.31YY43 pKa = 10.82VKK45 pKa = 10.35APRR48 pKa = 11.84AIRR51 pKa = 11.84PSKK54 pKa = 10.26STTKK58 pKa = 10.73SLGGTIGAHH67 pKa = 6.52LGHH70 pKa = 7.14GLQTVIKK77 pKa = 9.03ALTGFGDD84 pKa = 4.16YY85 pKa = 10.26EE86 pKa = 4.37VEE88 pKa = 4.64SNSLMPGTLGGDD100 pKa = 3.31PPIIQNSRR108 pKa = 11.84NNSHH112 pKa = 5.81VVHH115 pKa = 6.13HH116 pKa = 6.81RR117 pKa = 11.84EE118 pKa = 4.18YY119 pKa = 10.63IGDD122 pKa = 4.03VYY124 pKa = 11.41ASTAFSIQTYY134 pKa = 9.07PLNPGLLSSFPWVSQQADD152 pKa = 3.33SYY154 pKa = 9.87EE155 pKa = 4.03QYY157 pKa = 10.88KK158 pKa = 10.21FRR160 pKa = 11.84GAVFEE165 pKa = 4.31FKK167 pKa = 10.93SMSSDD172 pKa = 3.28AVLSSAASSSLGTVIMSTQYY192 pKa = 11.19NALEE196 pKa = 4.49APFSDD201 pKa = 3.04KK202 pKa = 10.37RR203 pKa = 11.84TMEE206 pKa = 3.93NYY208 pKa = 10.07EE209 pKa = 4.08YY210 pKa = 11.19ANSAKK215 pKa = 9.84PSCSMLHH222 pKa = 6.55PIEE225 pKa = 5.28CKK227 pKa = 9.59MSQTTVSEE235 pKa = 4.54LYY237 pKa = 10.24VRR239 pKa = 11.84TGVVSQGDD247 pKa = 3.83LRR249 pKa = 11.84LFDD252 pKa = 5.0LGNFSIAVQGKK263 pKa = 7.98QNVGANQVIGEE274 pKa = 4.33LGVV277 pKa = 3.23
MM1 pKa = 6.76STKK4 pKa = 10.24KK5 pKa = 9.31RR6 pKa = 11.84TAVPARR12 pKa = 11.84KK13 pKa = 7.38TVAAPKK19 pKa = 8.84RR20 pKa = 11.84TYY22 pKa = 11.1AKK24 pKa = 8.92RR25 pKa = 11.84TYY27 pKa = 7.88TARR30 pKa = 11.84APAATRR36 pKa = 11.84YY37 pKa = 9.79RR38 pKa = 11.84GTGDD42 pKa = 3.31YY43 pKa = 10.82VKK45 pKa = 10.35APRR48 pKa = 11.84AIRR51 pKa = 11.84PSKK54 pKa = 10.26STTKK58 pKa = 10.73SLGGTIGAHH67 pKa = 6.52LGHH70 pKa = 7.14GLQTVIKK77 pKa = 9.03ALTGFGDD84 pKa = 4.16YY85 pKa = 10.26EE86 pKa = 4.37VEE88 pKa = 4.64SNSLMPGTLGGDD100 pKa = 3.31PPIIQNSRR108 pKa = 11.84NNSHH112 pKa = 5.81VVHH115 pKa = 6.13HH116 pKa = 6.81RR117 pKa = 11.84EE118 pKa = 4.18YY119 pKa = 10.63IGDD122 pKa = 4.03VYY124 pKa = 11.41ASTAFSIQTYY134 pKa = 9.07PLNPGLLSSFPWVSQQADD152 pKa = 3.33SYY154 pKa = 9.87EE155 pKa = 4.03QYY157 pKa = 10.88KK158 pKa = 10.21FRR160 pKa = 11.84GAVFEE165 pKa = 4.31FKK167 pKa = 10.93SMSSDD172 pKa = 3.28AVLSSAASSSLGTVIMSTQYY192 pKa = 11.19NALEE196 pKa = 4.49APFSDD201 pKa = 3.04KK202 pKa = 10.37RR203 pKa = 11.84TMEE206 pKa = 3.93NYY208 pKa = 10.07EE209 pKa = 4.08YY210 pKa = 11.19ANSAKK215 pKa = 9.84PSCSMLHH222 pKa = 6.55PIEE225 pKa = 5.28CKK227 pKa = 9.59MSQTTVSEE235 pKa = 4.54LYY237 pKa = 10.24VRR239 pKa = 11.84TGVVSQGDD247 pKa = 3.83LRR249 pKa = 11.84LFDD252 pKa = 5.0LGNFSIAVQGKK263 pKa = 7.98QNVGANQVIGEE274 pKa = 4.33LGVV277 pKa = 3.23
Molecular weight: 29.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
643 |
277 |
366 |
321.5 |
36.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.02 ± 0.225 | 1.866 ± 0.703 |
4.821 ± 0.966 | 5.443 ± 0.904 |
3.733 ± 0.297 | 6.065 ± 1.375 |
1.866 ± 0.184 | 4.51 ± 0.331 |
7.621 ± 1.355 | 7.465 ± 0.594 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.333 ± 0.119 | 4.355 ± 0.236 |
4.355 ± 0.43 | 3.11 ± 0.751 |
6.687 ± 0.782 | 9.02 ± 1.334 |
6.065 ± 1.153 | 6.065 ± 0.71 |
2.022 ± 1.02 | 3.577 ± 0.908 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
