
Macaca mulatta feces associated virus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus macas1
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
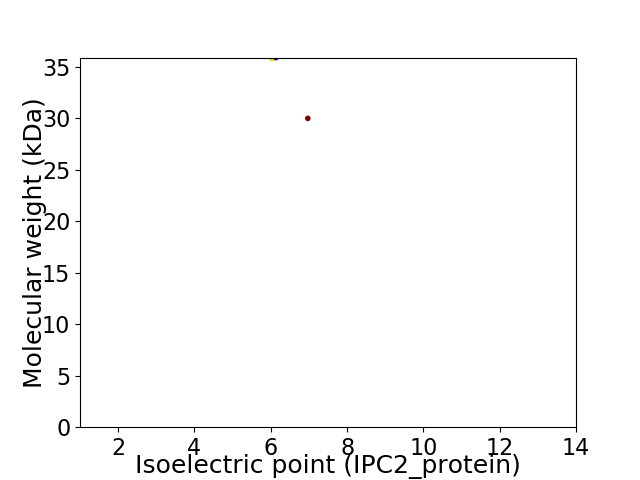
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5PZM4|A0A1W5PZM4_9VIRU Rep OS=Macaca mulatta feces associated virus 7 OX=2499229 PE=4 SV=1
MM1 pKa = 7.16IMKK4 pKa = 9.91VKK6 pKa = 9.17FTQFFDD12 pKa = 3.15ISSDD16 pKa = 3.3STKK19 pKa = 10.19FSKK22 pKa = 9.84FTVTAGGDD30 pKa = 3.78YY31 pKa = 11.21VKK33 pKa = 10.05MRR35 pKa = 11.84CAPQFVCYY43 pKa = 9.94KK44 pKa = 9.13YY45 pKa = 10.79FKK47 pKa = 10.34LGPVSVKK54 pKa = 10.28LVPASTLPVDD64 pKa = 3.87PTGLSYY70 pKa = 9.86EE71 pKa = 4.42TGEE74 pKa = 4.39STVDD78 pKa = 3.19PRR80 pKa = 11.84DD81 pKa = 3.38MFNPGLVRR89 pKa = 11.84ITNGEE94 pKa = 3.87PVGDD98 pKa = 4.77FNASDD103 pKa = 3.49MNQYY107 pKa = 10.5YY108 pKa = 8.07ATLLDD113 pKa = 3.72PRR115 pKa = 11.84WFKK118 pKa = 10.86FQLQQGFSRR127 pKa = 11.84YY128 pKa = 9.57AKK130 pKa = 9.5PLVYY134 pKa = 10.6NIGEE138 pKa = 4.18IAQEE142 pKa = 4.03PFPFAGTHH150 pKa = 5.46ISVSNNVSPEE160 pKa = 3.84YY161 pKa = 10.6YY162 pKa = 9.66YY163 pKa = 10.59QQSDD167 pKa = 3.74GSFKK171 pKa = 11.0SGGSAADD178 pKa = 3.38TEE180 pKa = 4.93RR181 pKa = 11.84YY182 pKa = 9.56IDD184 pKa = 3.5THH186 pKa = 6.35TPLQTGRR193 pKa = 11.84VKK195 pKa = 10.73LGWMPTDD202 pKa = 3.35SVMKK206 pKa = 9.69MATAAVNGVNTFSPNAAAYY225 pKa = 10.05VPEE228 pKa = 5.5IDD230 pKa = 5.5LITFILPKK238 pKa = 10.39AFKK241 pKa = 9.56TKK243 pKa = 9.26FFYY246 pKa = 10.5RR247 pKa = 11.84VFVEE251 pKa = 3.88STVYY255 pKa = 10.27FKK257 pKa = 11.32EE258 pKa = 4.09PVAVNPIIEE267 pKa = 4.63RR268 pKa = 11.84DD269 pKa = 3.6TDD271 pKa = 3.41KK272 pKa = 11.38SLYY275 pKa = 10.53AYY277 pKa = 10.4LPLDD281 pKa = 4.08RR282 pKa = 11.84GVGMQIKK289 pKa = 10.16DD290 pKa = 2.98IRR292 pKa = 11.84SYY294 pKa = 11.57TNAQKK299 pKa = 10.53IPLGDD304 pKa = 3.43SLGYY308 pKa = 9.73HH309 pKa = 6.14SKK311 pKa = 10.54VDD313 pKa = 3.82SVTGGAEE320 pKa = 3.82EE321 pKa = 4.01
MM1 pKa = 7.16IMKK4 pKa = 9.91VKK6 pKa = 9.17FTQFFDD12 pKa = 3.15ISSDD16 pKa = 3.3STKK19 pKa = 10.19FSKK22 pKa = 9.84FTVTAGGDD30 pKa = 3.78YY31 pKa = 11.21VKK33 pKa = 10.05MRR35 pKa = 11.84CAPQFVCYY43 pKa = 9.94KK44 pKa = 9.13YY45 pKa = 10.79FKK47 pKa = 10.34LGPVSVKK54 pKa = 10.28LVPASTLPVDD64 pKa = 3.87PTGLSYY70 pKa = 9.86EE71 pKa = 4.42TGEE74 pKa = 4.39STVDD78 pKa = 3.19PRR80 pKa = 11.84DD81 pKa = 3.38MFNPGLVRR89 pKa = 11.84ITNGEE94 pKa = 3.87PVGDD98 pKa = 4.77FNASDD103 pKa = 3.49MNQYY107 pKa = 10.5YY108 pKa = 8.07ATLLDD113 pKa = 3.72PRR115 pKa = 11.84WFKK118 pKa = 10.86FQLQQGFSRR127 pKa = 11.84YY128 pKa = 9.57AKK130 pKa = 9.5PLVYY134 pKa = 10.6NIGEE138 pKa = 4.18IAQEE142 pKa = 4.03PFPFAGTHH150 pKa = 5.46ISVSNNVSPEE160 pKa = 3.84YY161 pKa = 10.6YY162 pKa = 9.66YY163 pKa = 10.59QQSDD167 pKa = 3.74GSFKK171 pKa = 11.0SGGSAADD178 pKa = 3.38TEE180 pKa = 4.93RR181 pKa = 11.84YY182 pKa = 9.56IDD184 pKa = 3.5THH186 pKa = 6.35TPLQTGRR193 pKa = 11.84VKK195 pKa = 10.73LGWMPTDD202 pKa = 3.35SVMKK206 pKa = 9.69MATAAVNGVNTFSPNAAAYY225 pKa = 10.05VPEE228 pKa = 5.5IDD230 pKa = 5.5LITFILPKK238 pKa = 10.39AFKK241 pKa = 9.56TKK243 pKa = 9.26FFYY246 pKa = 10.5RR247 pKa = 11.84VFVEE251 pKa = 3.88STVYY255 pKa = 10.27FKK257 pKa = 11.32EE258 pKa = 4.09PVAVNPIIEE267 pKa = 4.63RR268 pKa = 11.84DD269 pKa = 3.6TDD271 pKa = 3.41KK272 pKa = 11.38SLYY275 pKa = 10.53AYY277 pKa = 10.4LPLDD281 pKa = 4.08RR282 pKa = 11.84GVGMQIKK289 pKa = 10.16DD290 pKa = 2.98IRR292 pKa = 11.84SYY294 pKa = 11.57TNAQKK299 pKa = 10.53IPLGDD304 pKa = 3.43SLGYY308 pKa = 9.73HH309 pKa = 6.14SKK311 pKa = 10.54VDD313 pKa = 3.82SVTGGAEE320 pKa = 3.82EE321 pKa = 4.01
Molecular weight: 35.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5PZM4|A0A1W5PZM4_9VIRU Rep OS=Macaca mulatta feces associated virus 7 OX=2499229 PE=4 SV=1
MM1 pKa = 7.22TNKK4 pKa = 9.64IYY6 pKa = 10.18MITMPKK12 pKa = 9.9NGRR15 pKa = 11.84NKK17 pKa = 10.4YY18 pKa = 9.57LLKK21 pKa = 10.92NLIEE25 pKa = 4.45TNDD28 pKa = 3.57LHH30 pKa = 6.73KK31 pKa = 10.07WAYY34 pKa = 9.99GYY36 pKa = 10.55EE37 pKa = 3.97IGKK40 pKa = 9.59NGYY43 pKa = 8.48KK44 pKa = 10.22HH45 pKa = 4.57IQARR49 pKa = 11.84IRR51 pKa = 11.84CPFDD55 pKa = 3.77FDD57 pKa = 4.0YY58 pKa = 11.1LQEE61 pKa = 4.3YY62 pKa = 9.5FGQAHH67 pKa = 6.49IEE69 pKa = 4.09EE70 pKa = 5.36ASDD73 pKa = 2.99EE74 pKa = 4.01WDD76 pKa = 3.51YY77 pKa = 10.24EE78 pKa = 4.68TKK80 pKa = 10.72GGIYY84 pKa = 8.13FTSEE88 pKa = 3.37DD89 pKa = 2.84WGEE92 pKa = 4.01RR93 pKa = 11.84LKK95 pKa = 10.81QRR97 pKa = 11.84YY98 pKa = 8.46LPLKK102 pKa = 9.36EE103 pKa = 4.29VQNKK107 pKa = 9.52ALEE110 pKa = 4.35GLEE113 pKa = 3.97ATNDD117 pKa = 3.36RR118 pKa = 11.84EE119 pKa = 4.33VYY121 pKa = 9.31VWYY124 pKa = 9.85DD125 pKa = 3.32EE126 pKa = 4.51KK127 pKa = 11.79GNAGKK132 pKa = 10.08SWLCGHH138 pKa = 6.98LWEE141 pKa = 4.71TGKK144 pKa = 10.88AYY146 pKa = 10.72VVDD149 pKa = 3.94SDD151 pKa = 4.01KK152 pKa = 11.89GEE154 pKa = 4.27TIKK157 pKa = 10.89KK158 pKa = 10.08DD159 pKa = 3.39VANEE163 pKa = 4.12YY164 pKa = 10.09INHH167 pKa = 5.53GWRR170 pKa = 11.84PYY172 pKa = 10.95VIIDD176 pKa = 4.03LPRR179 pKa = 11.84TAKK182 pKa = 8.39WTDD185 pKa = 3.06EE186 pKa = 4.05LYY188 pKa = 11.12YY189 pKa = 10.63AIEE192 pKa = 4.52KK193 pKa = 10.47IKK195 pKa = 10.97DD196 pKa = 3.62GLLKK200 pKa = 10.6DD201 pKa = 3.9PRR203 pKa = 11.84YY204 pKa = 9.68QSKK207 pKa = 7.83TVNIHH212 pKa = 4.58GVKK215 pKa = 10.24VMVMCNHH222 pKa = 6.92RR223 pKa = 11.84PNVSKK228 pKa = 11.02LSADD232 pKa = 3.22RR233 pKa = 11.84WKK235 pKa = 10.43MFTGAAAPEE244 pKa = 4.2PPAMQTAGGLSRR256 pKa = 11.84SSS258 pKa = 3.15
MM1 pKa = 7.22TNKK4 pKa = 9.64IYY6 pKa = 10.18MITMPKK12 pKa = 9.9NGRR15 pKa = 11.84NKK17 pKa = 10.4YY18 pKa = 9.57LLKK21 pKa = 10.92NLIEE25 pKa = 4.45TNDD28 pKa = 3.57LHH30 pKa = 6.73KK31 pKa = 10.07WAYY34 pKa = 9.99GYY36 pKa = 10.55EE37 pKa = 3.97IGKK40 pKa = 9.59NGYY43 pKa = 8.48KK44 pKa = 10.22HH45 pKa = 4.57IQARR49 pKa = 11.84IRR51 pKa = 11.84CPFDD55 pKa = 3.77FDD57 pKa = 4.0YY58 pKa = 11.1LQEE61 pKa = 4.3YY62 pKa = 9.5FGQAHH67 pKa = 6.49IEE69 pKa = 4.09EE70 pKa = 5.36ASDD73 pKa = 2.99EE74 pKa = 4.01WDD76 pKa = 3.51YY77 pKa = 10.24EE78 pKa = 4.68TKK80 pKa = 10.72GGIYY84 pKa = 8.13FTSEE88 pKa = 3.37DD89 pKa = 2.84WGEE92 pKa = 4.01RR93 pKa = 11.84LKK95 pKa = 10.81QRR97 pKa = 11.84YY98 pKa = 8.46LPLKK102 pKa = 9.36EE103 pKa = 4.29VQNKK107 pKa = 9.52ALEE110 pKa = 4.35GLEE113 pKa = 3.97ATNDD117 pKa = 3.36RR118 pKa = 11.84EE119 pKa = 4.33VYY121 pKa = 9.31VWYY124 pKa = 9.85DD125 pKa = 3.32EE126 pKa = 4.51KK127 pKa = 11.79GNAGKK132 pKa = 10.08SWLCGHH138 pKa = 6.98LWEE141 pKa = 4.71TGKK144 pKa = 10.88AYY146 pKa = 10.72VVDD149 pKa = 3.94SDD151 pKa = 4.01KK152 pKa = 11.89GEE154 pKa = 4.27TIKK157 pKa = 10.89KK158 pKa = 10.08DD159 pKa = 3.39VANEE163 pKa = 4.12YY164 pKa = 10.09INHH167 pKa = 5.53GWRR170 pKa = 11.84PYY172 pKa = 10.95VIIDD176 pKa = 4.03LPRR179 pKa = 11.84TAKK182 pKa = 8.39WTDD185 pKa = 3.06EE186 pKa = 4.05LYY188 pKa = 11.12YY189 pKa = 10.63AIEE192 pKa = 4.52KK193 pKa = 10.47IKK195 pKa = 10.97DD196 pKa = 3.62GLLKK200 pKa = 10.6DD201 pKa = 3.9PRR203 pKa = 11.84YY204 pKa = 9.68QSKK207 pKa = 7.83TVNIHH212 pKa = 4.58GVKK215 pKa = 10.24VMVMCNHH222 pKa = 6.92RR223 pKa = 11.84PNVSKK228 pKa = 11.02LSADD232 pKa = 3.22RR233 pKa = 11.84WKK235 pKa = 10.43MFTGAAAPEE244 pKa = 4.2PPAMQTAGGLSRR256 pKa = 11.84SSS258 pKa = 3.15
Molecular weight: 29.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
579 |
258 |
321 |
289.5 |
32.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.563 ± 0.017 | 0.864 ± 0.191 |
6.218 ± 0.01 | 5.699 ± 1.309 |
4.663 ± 1.739 | 7.427 ± 0.208 |
1.727 ± 0.629 | 5.181 ± 0.404 |
7.945 ± 1.113 | 6.218 ± 0.484 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.763 ± 0.032 | 4.491 ± 0.597 |
5.527 ± 1.053 | 3.282 ± 0.363 |
3.972 ± 0.433 | 6.045 ± 1.384 |
6.39 ± 0.862 | 6.736 ± 1.33 |
1.9 ± 1.013 | 6.39 ± 0.374 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
