
Circoviridae 8 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 8.39
Get precalculated fractions of proteins
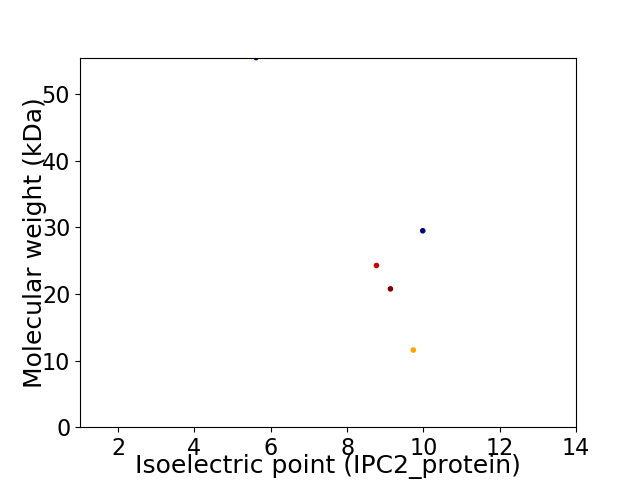
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|S5TND2|S5TND2_9CIRC Uncharacterized protein OS=Circoviridae 8 LDMD-2013 OX=1379712 PE=4 SV=1
MM1 pKa = 8.01IPICHH6 pKa = 6.92KK7 pKa = 10.52GGCGSFWLTKK17 pKa = 10.32KK18 pKa = 10.49KK19 pKa = 10.73LEE21 pKa = 3.93RR22 pKa = 11.84TEE24 pKa = 4.54RR25 pKa = 11.84CICRR29 pKa = 11.84VMCTLKK35 pKa = 8.4MTKK38 pKa = 9.16TFSTVRR44 pKa = 11.84NLCPQAHH51 pKa = 6.06WEE53 pKa = 4.0PRR55 pKa = 11.84KK56 pKa = 10.19GSHH59 pKa = 5.0QQAVDD64 pKa = 3.51YY65 pKa = 7.47CTKK68 pKa = 10.71EE69 pKa = 3.66EE70 pKa = 4.0TRR72 pKa = 11.84RR73 pKa = 11.84PGCDD77 pKa = 2.66PVVVGDD83 pKa = 4.64PPSQGKK89 pKa = 7.91RR90 pKa = 11.84TDD92 pKa = 3.08IMDD95 pKa = 3.45VKK97 pKa = 11.09SFIDD101 pKa = 3.95GGCSRR106 pKa = 11.84EE107 pKa = 4.18EE108 pKa = 3.61LWSEE112 pKa = 4.01HH113 pKa = 5.31FSAMSKK119 pKa = 9.39MYY121 pKa = 10.48KK122 pKa = 10.42AMYY125 pKa = 8.71EE126 pKa = 4.0YY127 pKa = 11.12SFVRR131 pKa = 11.84SGEE134 pKa = 4.38RR135 pKa = 11.84NWKK138 pKa = 7.92TRR140 pKa = 11.84VTVIVGPPGTGKK152 pKa = 11.22SMFCNNYY159 pKa = 9.54HH160 pKa = 5.98GQSYY164 pKa = 8.27WKK166 pKa = 10.4NNSLFFDD173 pKa = 5.37GYY175 pKa = 10.23HH176 pKa = 5.44GQDD179 pKa = 3.96VIVMDD184 pKa = 4.83DD185 pKa = 4.57FYY187 pKa = 11.8GQLPWSTLLKK197 pKa = 10.63LADD200 pKa = 4.4RR201 pKa = 11.84YY202 pKa = 11.18KK203 pKa = 11.07LILDD207 pKa = 3.78IKK209 pKa = 9.45GSSVNMLAKK218 pKa = 10.3EE219 pKa = 4.03LFITSNADD227 pKa = 2.86IEE229 pKa = 4.35EE230 pKa = 4.51WYY232 pKa = 10.29SYY234 pKa = 10.62KK235 pKa = 10.54PGMDD239 pKa = 2.51IRR241 pKa = 11.84ALEE244 pKa = 3.92RR245 pKa = 11.84RR246 pKa = 11.84IDD248 pKa = 3.4HH249 pKa = 5.81YY250 pKa = 12.0VNVDD254 pKa = 3.25EE255 pKa = 6.14SGTITNRR262 pKa = 11.84SSYY265 pKa = 9.85EE266 pKa = 3.46PHH268 pKa = 7.49KK269 pKa = 10.86ILTNGKK275 pKa = 9.48KK276 pKa = 10.21LVDD279 pKa = 3.61MANLIKK285 pKa = 10.98NFVFSYY291 pKa = 10.33KK292 pKa = 10.05PSEE295 pKa = 4.68DD296 pKa = 3.2GSGFSTNVTIDD307 pKa = 3.03EE308 pKa = 4.43YY309 pKa = 10.79EE310 pKa = 3.93IRR312 pKa = 11.84RR313 pKa = 11.84HH314 pKa = 4.76LTVNPEE320 pKa = 3.91VLSAPEE326 pKa = 3.94FLEE329 pKa = 5.34LSDD332 pKa = 4.94KK333 pKa = 10.99LSHH336 pKa = 6.23TLNEE340 pKa = 4.09LEE342 pKa = 4.26YY343 pKa = 10.69QVAEE347 pKa = 4.35LRR349 pKa = 11.84RR350 pKa = 11.84KK351 pKa = 8.58VANEE355 pKa = 3.77LVDD358 pKa = 3.81VPINFQRR365 pKa = 11.84EE366 pKa = 4.18SSSNLSARR374 pKa = 11.84DD375 pKa = 3.23IMEE378 pKa = 3.93VAEE381 pKa = 4.2RR382 pKa = 11.84AVEE385 pKa = 4.11EE386 pKa = 4.49VRR388 pKa = 11.84GHH390 pKa = 6.86PNLEE394 pKa = 4.04LEE396 pKa = 4.86DD397 pKa = 3.61RR398 pKa = 11.84HH399 pKa = 5.77HH400 pKa = 7.16TYY402 pKa = 8.5GTPTPRR408 pKa = 11.84KK409 pKa = 9.26RR410 pKa = 11.84RR411 pKa = 11.84RR412 pKa = 11.84DD413 pKa = 3.52EE414 pKa = 4.27PNISPRR420 pKa = 11.84SRR422 pKa = 11.84RR423 pKa = 11.84PPRR426 pKa = 11.84IGRR429 pKa = 11.84SGGEE433 pKa = 3.87NIIPMRR439 pKa = 11.84DD440 pKa = 3.31VNEE443 pKa = 4.4LSNSSEE449 pKa = 3.85GEE451 pKa = 4.17SFNTDD456 pKa = 2.51EE457 pKa = 5.85DD458 pKa = 3.8EE459 pKa = 6.12AEE461 pKa = 4.19MYY463 pKa = 10.7EE464 pKa = 4.22EE465 pKa = 4.75GLNTQHH471 pKa = 6.7TSEE474 pKa = 4.27TLGTEE479 pKa = 4.34DD480 pKa = 4.04TMEE483 pKa = 4.01
MM1 pKa = 8.01IPICHH6 pKa = 6.92KK7 pKa = 10.52GGCGSFWLTKK17 pKa = 10.32KK18 pKa = 10.49KK19 pKa = 10.73LEE21 pKa = 3.93RR22 pKa = 11.84TEE24 pKa = 4.54RR25 pKa = 11.84CICRR29 pKa = 11.84VMCTLKK35 pKa = 8.4MTKK38 pKa = 9.16TFSTVRR44 pKa = 11.84NLCPQAHH51 pKa = 6.06WEE53 pKa = 4.0PRR55 pKa = 11.84KK56 pKa = 10.19GSHH59 pKa = 5.0QQAVDD64 pKa = 3.51YY65 pKa = 7.47CTKK68 pKa = 10.71EE69 pKa = 3.66EE70 pKa = 4.0TRR72 pKa = 11.84RR73 pKa = 11.84PGCDD77 pKa = 2.66PVVVGDD83 pKa = 4.64PPSQGKK89 pKa = 7.91RR90 pKa = 11.84TDD92 pKa = 3.08IMDD95 pKa = 3.45VKK97 pKa = 11.09SFIDD101 pKa = 3.95GGCSRR106 pKa = 11.84EE107 pKa = 4.18EE108 pKa = 3.61LWSEE112 pKa = 4.01HH113 pKa = 5.31FSAMSKK119 pKa = 9.39MYY121 pKa = 10.48KK122 pKa = 10.42AMYY125 pKa = 8.71EE126 pKa = 4.0YY127 pKa = 11.12SFVRR131 pKa = 11.84SGEE134 pKa = 4.38RR135 pKa = 11.84NWKK138 pKa = 7.92TRR140 pKa = 11.84VTVIVGPPGTGKK152 pKa = 11.22SMFCNNYY159 pKa = 9.54HH160 pKa = 5.98GQSYY164 pKa = 8.27WKK166 pKa = 10.4NNSLFFDD173 pKa = 5.37GYY175 pKa = 10.23HH176 pKa = 5.44GQDD179 pKa = 3.96VIVMDD184 pKa = 4.83DD185 pKa = 4.57FYY187 pKa = 11.8GQLPWSTLLKK197 pKa = 10.63LADD200 pKa = 4.4RR201 pKa = 11.84YY202 pKa = 11.18KK203 pKa = 11.07LILDD207 pKa = 3.78IKK209 pKa = 9.45GSSVNMLAKK218 pKa = 10.3EE219 pKa = 4.03LFITSNADD227 pKa = 2.86IEE229 pKa = 4.35EE230 pKa = 4.51WYY232 pKa = 10.29SYY234 pKa = 10.62KK235 pKa = 10.54PGMDD239 pKa = 2.51IRR241 pKa = 11.84ALEE244 pKa = 3.92RR245 pKa = 11.84RR246 pKa = 11.84IDD248 pKa = 3.4HH249 pKa = 5.81YY250 pKa = 12.0VNVDD254 pKa = 3.25EE255 pKa = 6.14SGTITNRR262 pKa = 11.84SSYY265 pKa = 9.85EE266 pKa = 3.46PHH268 pKa = 7.49KK269 pKa = 10.86ILTNGKK275 pKa = 9.48KK276 pKa = 10.21LVDD279 pKa = 3.61MANLIKK285 pKa = 10.98NFVFSYY291 pKa = 10.33KK292 pKa = 10.05PSEE295 pKa = 4.68DD296 pKa = 3.2GSGFSTNVTIDD307 pKa = 3.03EE308 pKa = 4.43YY309 pKa = 10.79EE310 pKa = 3.93IRR312 pKa = 11.84RR313 pKa = 11.84HH314 pKa = 4.76LTVNPEE320 pKa = 3.91VLSAPEE326 pKa = 3.94FLEE329 pKa = 5.34LSDD332 pKa = 4.94KK333 pKa = 10.99LSHH336 pKa = 6.23TLNEE340 pKa = 4.09LEE342 pKa = 4.26YY343 pKa = 10.69QVAEE347 pKa = 4.35LRR349 pKa = 11.84RR350 pKa = 11.84KK351 pKa = 8.58VANEE355 pKa = 3.77LVDD358 pKa = 3.81VPINFQRR365 pKa = 11.84EE366 pKa = 4.18SSSNLSARR374 pKa = 11.84DD375 pKa = 3.23IMEE378 pKa = 3.93VAEE381 pKa = 4.2RR382 pKa = 11.84AVEE385 pKa = 4.11EE386 pKa = 4.49VRR388 pKa = 11.84GHH390 pKa = 6.86PNLEE394 pKa = 4.04LEE396 pKa = 4.86DD397 pKa = 3.61RR398 pKa = 11.84HH399 pKa = 5.77HH400 pKa = 7.16TYY402 pKa = 8.5GTPTPRR408 pKa = 11.84KK409 pKa = 9.26RR410 pKa = 11.84RR411 pKa = 11.84RR412 pKa = 11.84DD413 pKa = 3.52EE414 pKa = 4.27PNISPRR420 pKa = 11.84SRR422 pKa = 11.84RR423 pKa = 11.84PPRR426 pKa = 11.84IGRR429 pKa = 11.84SGGEE433 pKa = 3.87NIIPMRR439 pKa = 11.84DD440 pKa = 3.31VNEE443 pKa = 4.4LSNSSEE449 pKa = 3.85GEE451 pKa = 4.17SFNTDD456 pKa = 2.51EE457 pKa = 5.85DD458 pKa = 3.8EE459 pKa = 6.12AEE461 pKa = 4.19MYY463 pKa = 10.7EE464 pKa = 4.22EE465 pKa = 4.75GLNTQHH471 pKa = 6.7TSEE474 pKa = 4.27TLGTEE479 pKa = 4.34DD480 pKa = 4.04TMEE483 pKa = 4.01
Molecular weight: 55.45 kDa
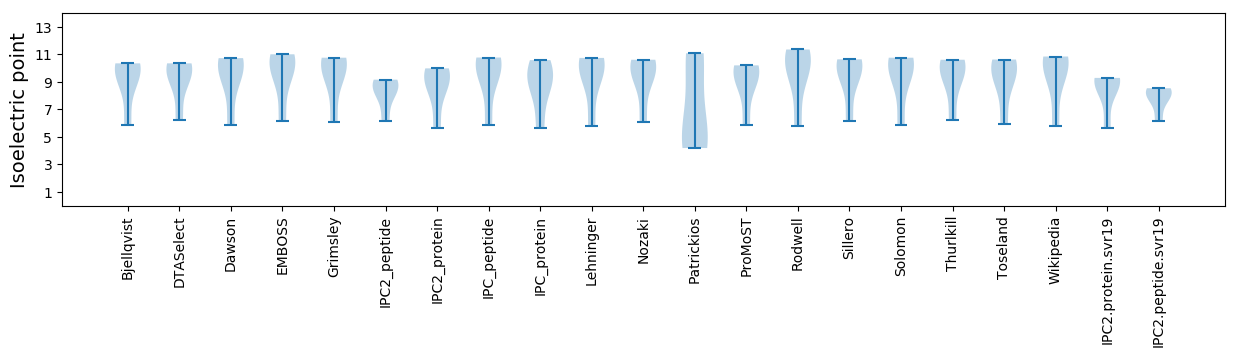
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5SYD5|S5SYD5_9CIRC Putative viral capsid OS=Circoviridae 8 LDMD-2013 OX=1379712 PE=4 SV=1
MM1 pKa = 7.85GKK3 pKa = 9.66RR4 pKa = 11.84KK5 pKa = 9.66SRR7 pKa = 11.84SRR9 pKa = 11.84QTSISKK15 pKa = 8.41YY16 pKa = 6.68VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84TNFSQTPQPPPLYY34 pKa = 10.28HH35 pKa = 6.44RR36 pKa = 11.84SKK38 pKa = 10.22WKK40 pKa = 9.87YY41 pKa = 9.3AGNMWVRR48 pKa = 11.84KK49 pKa = 9.38NKK51 pKa = 10.64GEE53 pKa = 3.95IHH55 pKa = 6.86FDD57 pKa = 3.04RR58 pKa = 11.84YY59 pKa = 10.74GYY61 pKa = 9.28PLPSAVNRR69 pKa = 11.84EE70 pKa = 4.08EE71 pKa = 4.08YY72 pKa = 10.61KK73 pKa = 10.61DD74 pKa = 3.44WKK76 pKa = 10.14APYY79 pKa = 9.71YY80 pKa = 10.13PWDD83 pKa = 4.22EE84 pKa = 3.98KK85 pKa = 10.96WKK87 pKa = 7.17TTSYY91 pKa = 10.94TNAGYY96 pKa = 10.75AWGTPIYY103 pKa = 10.24DD104 pKa = 3.95DD105 pKa = 3.96QRR107 pKa = 11.84LMGHH111 pKa = 5.66KK112 pKa = 9.69VRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84DD117 pKa = 3.32GLYY120 pKa = 10.71EE121 pKa = 4.19EE122 pKa = 5.98LPADD126 pKa = 3.46WVLPPTTRR134 pKa = 11.84PTMTRR139 pKa = 11.84LQNTLFDD146 pKa = 4.15KK147 pKa = 10.52TYY149 pKa = 8.99HH150 pKa = 6.75PYY152 pKa = 10.27RR153 pKa = 11.84YY154 pKa = 9.82GYY156 pKa = 8.53TRR158 pKa = 11.84ALDD161 pKa = 3.11WARR164 pKa = 11.84RR165 pKa = 11.84HH166 pKa = 6.46RR167 pKa = 11.84IAVWIIKK174 pKa = 8.99RR175 pKa = 11.84IWWGFVLKK183 pKa = 10.88NGIYY187 pKa = 8.31PTEE190 pKa = 4.84DD191 pKa = 2.81FLQRR195 pKa = 11.84QVEE198 pKa = 4.2KK199 pKa = 10.55FKK201 pKa = 10.97RR202 pKa = 11.84YY203 pKa = 7.48MQGKK207 pKa = 8.48VSSSEE212 pKa = 3.61ADD214 pKa = 3.14PVVKK218 pKa = 10.57NPPQRR223 pKa = 11.84PIHH226 pKa = 5.96PSQRR230 pKa = 11.84PSKK233 pKa = 9.66IRR235 pKa = 11.84SRR237 pKa = 11.84IKK239 pKa = 10.21ANYY242 pKa = 8.02YY243 pKa = 10.16
MM1 pKa = 7.85GKK3 pKa = 9.66RR4 pKa = 11.84KK5 pKa = 9.66SRR7 pKa = 11.84SRR9 pKa = 11.84QTSISKK15 pKa = 8.41YY16 pKa = 6.68VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84TNFSQTPQPPPLYY34 pKa = 10.28HH35 pKa = 6.44RR36 pKa = 11.84SKK38 pKa = 10.22WKK40 pKa = 9.87YY41 pKa = 9.3AGNMWVRR48 pKa = 11.84KK49 pKa = 9.38NKK51 pKa = 10.64GEE53 pKa = 3.95IHH55 pKa = 6.86FDD57 pKa = 3.04RR58 pKa = 11.84YY59 pKa = 10.74GYY61 pKa = 9.28PLPSAVNRR69 pKa = 11.84EE70 pKa = 4.08EE71 pKa = 4.08YY72 pKa = 10.61KK73 pKa = 10.61DD74 pKa = 3.44WKK76 pKa = 10.14APYY79 pKa = 9.71YY80 pKa = 10.13PWDD83 pKa = 4.22EE84 pKa = 3.98KK85 pKa = 10.96WKK87 pKa = 7.17TTSYY91 pKa = 10.94TNAGYY96 pKa = 10.75AWGTPIYY103 pKa = 10.24DD104 pKa = 3.95DD105 pKa = 3.96QRR107 pKa = 11.84LMGHH111 pKa = 5.66KK112 pKa = 9.69VRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84DD117 pKa = 3.32GLYY120 pKa = 10.71EE121 pKa = 4.19EE122 pKa = 5.98LPADD126 pKa = 3.46WVLPPTTRR134 pKa = 11.84PTMTRR139 pKa = 11.84LQNTLFDD146 pKa = 4.15KK147 pKa = 10.52TYY149 pKa = 8.99HH150 pKa = 6.75PYY152 pKa = 10.27RR153 pKa = 11.84YY154 pKa = 9.82GYY156 pKa = 8.53TRR158 pKa = 11.84ALDD161 pKa = 3.11WARR164 pKa = 11.84RR165 pKa = 11.84HH166 pKa = 6.46RR167 pKa = 11.84IAVWIIKK174 pKa = 8.99RR175 pKa = 11.84IWWGFVLKK183 pKa = 10.88NGIYY187 pKa = 8.31PTEE190 pKa = 4.84DD191 pKa = 2.81FLQRR195 pKa = 11.84QVEE198 pKa = 4.2KK199 pKa = 10.55FKK201 pKa = 10.97RR202 pKa = 11.84YY203 pKa = 7.48MQGKK207 pKa = 8.48VSSSEE212 pKa = 3.61ADD214 pKa = 3.14PVVKK218 pKa = 10.57NPPQRR223 pKa = 11.84PIHH226 pKa = 5.96PSQRR230 pKa = 11.84PSKK233 pKa = 9.66IRR235 pKa = 11.84SRR237 pKa = 11.84IKK239 pKa = 10.21ANYY242 pKa = 8.02YY243 pKa = 10.16
Molecular weight: 29.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1228 |
102 |
483 |
245.6 |
28.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.479 ± 0.878 | 1.792 ± 0.505 |
4.479 ± 0.67 | 5.863 ± 1.567 |
3.502 ± 0.526 | 6.026 ± 0.537 |
3.583 ± 0.512 | 5.7 ± 0.54 |
7.248 ± 1.037 | 7.003 ± 1.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.199 ± 0.501 | 4.805 ± 0.334 |
6.27 ± 0.762 | 3.176 ± 0.487 |
7.736 ± 1.061 | 8.225 ± 1.01 |
5.863 ± 0.353 | 5.375 ± 0.453 |
1.954 ± 0.632 | 4.642 ± 0.914 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
