
Lake Sarah-associated circular virus-32
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.91
Get precalculated fractions of proteins
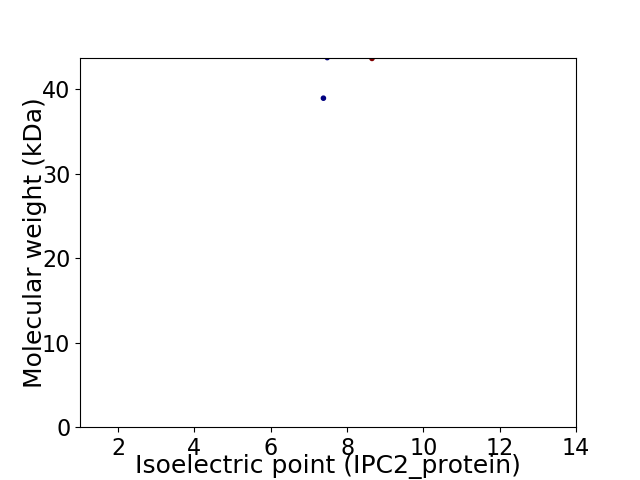
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
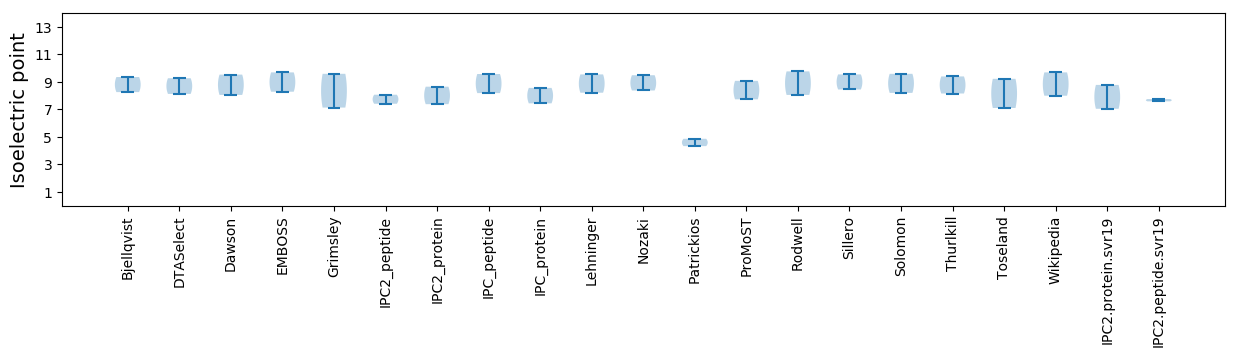
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GAA6|A0A126GAA6_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-32 OX=1685760 PE=3 SV=1
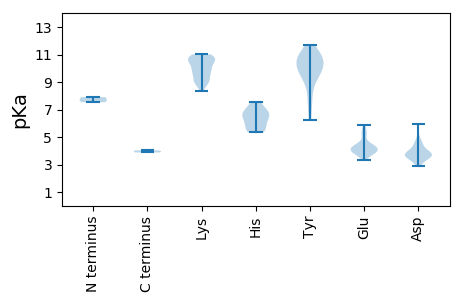
MM1 pKa = 7.53EE2 pKa = 5.37FDD4 pKa = 3.45KK5 pKa = 10.95EE6 pKa = 3.78FRR8 pKa = 11.84IPFRR12 pKa = 11.84QTTRR16 pKa = 11.84IVVTGEE22 pKa = 3.62AGNQYY27 pKa = 10.21FVLKK31 pKa = 10.51DD32 pKa = 3.27YY33 pKa = 11.06SRR35 pKa = 11.84WGAAIAADD43 pKa = 3.9TRR45 pKa = 11.84APTALGLLGQTDD57 pKa = 3.78TDD59 pKa = 4.48DD60 pKa = 4.72NCPQLQDD67 pKa = 3.72VYY69 pKa = 10.62SQLVNYY75 pKa = 7.34EE76 pKa = 3.82YY77 pKa = 11.2CKK79 pKa = 9.57IHH81 pKa = 6.61AVNEE85 pKa = 3.94EE86 pKa = 3.95FRR88 pKa = 11.84VVDD91 pKa = 3.47YY92 pKa = 9.93EE93 pKa = 4.24VRR95 pKa = 11.84GSGTTVALSRR105 pKa = 11.84NSPHH109 pKa = 5.94FTRR112 pKa = 11.84DD113 pKa = 2.98PSAQLLWKK121 pKa = 8.98LTTNGTEE128 pKa = 4.12EE129 pKa = 4.37SRR131 pKa = 11.84LLTAAAVDD139 pKa = 3.86GADD142 pKa = 3.8FMRR145 pKa = 11.84ALAHH149 pKa = 5.44TRR151 pKa = 11.84RR152 pKa = 11.84GTSFTTRR159 pKa = 11.84LPTEE163 pKa = 4.0MKK165 pKa = 9.82MMRR168 pKa = 11.84GQAGRR173 pKa = 11.84LLLSWTGRR181 pKa = 11.84EE182 pKa = 3.88ATPNITINGSILSNNANNLGPTGSSTASSAFNIRR216 pKa = 11.84NTSAGSGIVDD226 pKa = 4.61FSAHH230 pKa = 6.62RR231 pKa = 11.84SNIFYY236 pKa = 9.64WGKK239 pKa = 9.01GQIYY243 pKa = 10.61NMGGQCDD250 pKa = 3.81VNNYY254 pKa = 6.27QSKK257 pKa = 10.64YY258 pKa = 9.81YY259 pKa = 9.49MATATPYY266 pKa = 10.81AILPSNEE273 pKa = 3.36ARR275 pKa = 11.84NDD277 pKa = 2.88ISFPRR282 pKa = 11.84IFSCTPLIEE291 pKa = 4.44SVTAFNTEE299 pKa = 4.17RR300 pKa = 11.84VWKK303 pKa = 8.98IQNITTLIVKK313 pKa = 10.01ARR315 pKa = 11.84GYY317 pKa = 10.92VPTKK321 pKa = 10.42ALSSLKK327 pKa = 10.71FEE329 pKa = 4.38TQFLSVNDD337 pKa = 4.36DD338 pKa = 4.12DD339 pKa = 5.92EE340 pKa = 5.23PMSKK344 pKa = 9.99QIKK347 pKa = 10.01LIEE350 pKa = 4.03
MM1 pKa = 7.53EE2 pKa = 5.37FDD4 pKa = 3.45KK5 pKa = 10.95EE6 pKa = 3.78FRR8 pKa = 11.84IPFRR12 pKa = 11.84QTTRR16 pKa = 11.84IVVTGEE22 pKa = 3.62AGNQYY27 pKa = 10.21FVLKK31 pKa = 10.51DD32 pKa = 3.27YY33 pKa = 11.06SRR35 pKa = 11.84WGAAIAADD43 pKa = 3.9TRR45 pKa = 11.84APTALGLLGQTDD57 pKa = 3.78TDD59 pKa = 4.48DD60 pKa = 4.72NCPQLQDD67 pKa = 3.72VYY69 pKa = 10.62SQLVNYY75 pKa = 7.34EE76 pKa = 3.82YY77 pKa = 11.2CKK79 pKa = 9.57IHH81 pKa = 6.61AVNEE85 pKa = 3.94EE86 pKa = 3.95FRR88 pKa = 11.84VVDD91 pKa = 3.47YY92 pKa = 9.93EE93 pKa = 4.24VRR95 pKa = 11.84GSGTTVALSRR105 pKa = 11.84NSPHH109 pKa = 5.94FTRR112 pKa = 11.84DD113 pKa = 2.98PSAQLLWKK121 pKa = 8.98LTTNGTEE128 pKa = 4.12EE129 pKa = 4.37SRR131 pKa = 11.84LLTAAAVDD139 pKa = 3.86GADD142 pKa = 3.8FMRR145 pKa = 11.84ALAHH149 pKa = 5.44TRR151 pKa = 11.84RR152 pKa = 11.84GTSFTTRR159 pKa = 11.84LPTEE163 pKa = 4.0MKK165 pKa = 9.82MMRR168 pKa = 11.84GQAGRR173 pKa = 11.84LLLSWTGRR181 pKa = 11.84EE182 pKa = 3.88ATPNITINGSILSNNANNLGPTGSSTASSAFNIRR216 pKa = 11.84NTSAGSGIVDD226 pKa = 4.61FSAHH230 pKa = 6.62RR231 pKa = 11.84SNIFYY236 pKa = 9.64WGKK239 pKa = 9.01GQIYY243 pKa = 10.61NMGGQCDD250 pKa = 3.81VNNYY254 pKa = 6.27QSKK257 pKa = 10.64YY258 pKa = 9.81YY259 pKa = 9.49MATATPYY266 pKa = 10.81AILPSNEE273 pKa = 3.36ARR275 pKa = 11.84NDD277 pKa = 2.88ISFPRR282 pKa = 11.84IFSCTPLIEE291 pKa = 4.44SVTAFNTEE299 pKa = 4.17RR300 pKa = 11.84VWKK303 pKa = 8.98IQNITTLIVKK313 pKa = 10.01ARR315 pKa = 11.84GYY317 pKa = 10.92VPTKK321 pKa = 10.42ALSSLKK327 pKa = 10.71FEE329 pKa = 4.38TQFLSVNDD337 pKa = 4.36DD338 pKa = 4.12DD339 pKa = 5.92EE340 pKa = 5.23PMSKK344 pKa = 9.99QIKK347 pKa = 10.01LIEE350 pKa = 4.03
Molecular weight: 38.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GAA6|A0A126GAA6_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-32 OX=1685760 PE=3 SV=1
MM1 pKa = 7.9LIPIRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SLRR11 pKa = 11.84IHH13 pKa = 6.83LWIQPSAPPCGQSKK27 pKa = 8.84TIKK30 pKa = 9.75PRR32 pKa = 11.84PSQPKK37 pKa = 9.02LSVRR41 pKa = 11.84MTSFSSKK48 pKa = 9.12RR49 pKa = 11.84WCFTLNNYY57 pKa = 9.85SPEE60 pKa = 3.88EE61 pKa = 3.94HH62 pKa = 6.54QALQLAFKK70 pKa = 10.67ASTVKK75 pKa = 10.63YY76 pKa = 10.17AVIGIEE82 pKa = 4.17KK83 pKa = 10.04GEE85 pKa = 4.34SGTPHH90 pKa = 6.1LQGFVVFKK98 pKa = 10.85KK99 pKa = 8.33MQRR102 pKa = 11.84LSALKK107 pKa = 9.99KK108 pKa = 9.38LNPRR112 pKa = 11.84AHH114 pKa = 6.04YY115 pKa = 10.06EE116 pKa = 4.03EE117 pKa = 5.24AKK119 pKa = 9.65GTSQQASTYY128 pKa = 7.74CQKK131 pKa = 11.01EE132 pKa = 4.42GIFEE136 pKa = 4.22EE137 pKa = 5.56FGTCPLEE144 pKa = 4.04PTAIASNASNKK155 pKa = 9.87KK156 pKa = 8.99NEE158 pKa = 4.27QYY160 pKa = 10.73RR161 pKa = 11.84KK162 pKa = 10.72AMDD165 pKa = 3.79LAKK168 pKa = 10.81LGDD171 pKa = 4.12FSAIEE176 pKa = 4.04QEE178 pKa = 4.91WPGLWLRR185 pKa = 11.84HH186 pKa = 5.34RR187 pKa = 11.84NAFLSAPRR195 pKa = 11.84DD196 pKa = 3.52FGIRR200 pKa = 11.84PKK202 pKa = 11.0DD203 pKa = 3.71LDD205 pKa = 4.09YY206 pKa = 11.68LPGIWIWGPAGVGKK220 pKa = 10.45SRR222 pKa = 11.84LARR225 pKa = 11.84EE226 pKa = 4.27VFPLSYY232 pKa = 10.57EE233 pKa = 3.72KK234 pKa = 10.68RR235 pKa = 11.84LNKK238 pKa = 9.59WFDD241 pKa = 3.35GYY243 pKa = 10.8SAQKK247 pKa = 10.53AVLIEE252 pKa = 5.91DD253 pKa = 3.63IDD255 pKa = 3.61QSHH258 pKa = 6.88RR259 pKa = 11.84YY260 pKa = 8.83ISHH263 pKa = 7.52DD264 pKa = 3.55LKK266 pKa = 10.8IWTDD270 pKa = 3.37RR271 pKa = 11.84YY272 pKa = 10.97SFPAEE277 pKa = 3.76IKK279 pKa = 10.68GGAIQCRR286 pKa = 11.84PEE288 pKa = 3.95HH289 pKa = 6.55VLITSQYY296 pKa = 8.7HH297 pKa = 5.64IHH299 pKa = 7.54DD300 pKa = 3.56IWDD303 pKa = 3.8DD304 pKa = 3.72VQTRR308 pKa = 11.84EE309 pKa = 4.5AIGRR313 pKa = 11.84RR314 pKa = 11.84CFVIYY319 pKa = 9.81IDD321 pKa = 3.7KK322 pKa = 9.24ATPNAYY328 pKa = 9.41KK329 pKa = 10.64SSLSALNHH337 pKa = 6.93LIRR340 pKa = 11.84SRR342 pKa = 11.84APVGEE347 pKa = 4.35CTVVPVAPSDD357 pKa = 3.86VSASAHH363 pKa = 5.42VAPSPPVATLPVLFPDD379 pKa = 4.43PSDD382 pKa = 4.97LSVLDD387 pKa = 3.87
MM1 pKa = 7.9LIPIRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SLRR11 pKa = 11.84IHH13 pKa = 6.83LWIQPSAPPCGQSKK27 pKa = 8.84TIKK30 pKa = 9.75PRR32 pKa = 11.84PSQPKK37 pKa = 9.02LSVRR41 pKa = 11.84MTSFSSKK48 pKa = 9.12RR49 pKa = 11.84WCFTLNNYY57 pKa = 9.85SPEE60 pKa = 3.88EE61 pKa = 3.94HH62 pKa = 6.54QALQLAFKK70 pKa = 10.67ASTVKK75 pKa = 10.63YY76 pKa = 10.17AVIGIEE82 pKa = 4.17KK83 pKa = 10.04GEE85 pKa = 4.34SGTPHH90 pKa = 6.1LQGFVVFKK98 pKa = 10.85KK99 pKa = 8.33MQRR102 pKa = 11.84LSALKK107 pKa = 9.99KK108 pKa = 9.38LNPRR112 pKa = 11.84AHH114 pKa = 6.04YY115 pKa = 10.06EE116 pKa = 4.03EE117 pKa = 5.24AKK119 pKa = 9.65GTSQQASTYY128 pKa = 7.74CQKK131 pKa = 11.01EE132 pKa = 4.42GIFEE136 pKa = 4.22EE137 pKa = 5.56FGTCPLEE144 pKa = 4.04PTAIASNASNKK155 pKa = 9.87KK156 pKa = 8.99NEE158 pKa = 4.27QYY160 pKa = 10.73RR161 pKa = 11.84KK162 pKa = 10.72AMDD165 pKa = 3.79LAKK168 pKa = 10.81LGDD171 pKa = 4.12FSAIEE176 pKa = 4.04QEE178 pKa = 4.91WPGLWLRR185 pKa = 11.84HH186 pKa = 5.34RR187 pKa = 11.84NAFLSAPRR195 pKa = 11.84DD196 pKa = 3.52FGIRR200 pKa = 11.84PKK202 pKa = 11.0DD203 pKa = 3.71LDD205 pKa = 4.09YY206 pKa = 11.68LPGIWIWGPAGVGKK220 pKa = 10.45SRR222 pKa = 11.84LARR225 pKa = 11.84EE226 pKa = 4.27VFPLSYY232 pKa = 10.57EE233 pKa = 3.72KK234 pKa = 10.68RR235 pKa = 11.84LNKK238 pKa = 9.59WFDD241 pKa = 3.35GYY243 pKa = 10.8SAQKK247 pKa = 10.53AVLIEE252 pKa = 5.91DD253 pKa = 3.63IDD255 pKa = 3.61QSHH258 pKa = 6.88RR259 pKa = 11.84YY260 pKa = 8.83ISHH263 pKa = 7.52DD264 pKa = 3.55LKK266 pKa = 10.8IWTDD270 pKa = 3.37RR271 pKa = 11.84YY272 pKa = 10.97SFPAEE277 pKa = 3.76IKK279 pKa = 10.68GGAIQCRR286 pKa = 11.84PEE288 pKa = 3.95HH289 pKa = 6.55VLITSQYY296 pKa = 8.7HH297 pKa = 5.64IHH299 pKa = 7.54DD300 pKa = 3.56IWDD303 pKa = 3.8DD304 pKa = 3.72VQTRR308 pKa = 11.84EE309 pKa = 4.5AIGRR313 pKa = 11.84RR314 pKa = 11.84CFVIYY319 pKa = 9.81IDD321 pKa = 3.7KK322 pKa = 9.24ATPNAYY328 pKa = 9.41KK329 pKa = 10.64SSLSALNHH337 pKa = 6.93LIRR340 pKa = 11.84SRR342 pKa = 11.84APVGEE347 pKa = 4.35CTVVPVAPSDD357 pKa = 3.86VSASAHH363 pKa = 5.42VAPSPPVATLPVLFPDD379 pKa = 4.43PSDD382 pKa = 4.97LSVLDD387 pKa = 3.87
Molecular weight: 43.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
737 |
350 |
387 |
368.5 |
41.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.548 ± 0.015 | 1.493 ± 0.228 |
4.749 ± 0.071 | 5.02 ± 0.107 |
4.206 ± 0.239 | 5.834 ± 0.481 |
2.171 ± 0.672 | 6.242 ± 0.344 |
5.292 ± 1.031 | 7.87 ± 0.288 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.628 ± 0.43 | 4.478 ± 1.368 |
5.97 ± 1.287 | 4.206 ± 0.135 |
6.513 ± 0.038 | 8.548 ± 0.358 |
6.649 ± 2.003 | 5.156 ± 0.009 |
1.9 ± 0.308 | 3.528 ± 0.122 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
