
Capybara microvirus Cap1_SP_188
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.37
Get precalculated fractions of proteins
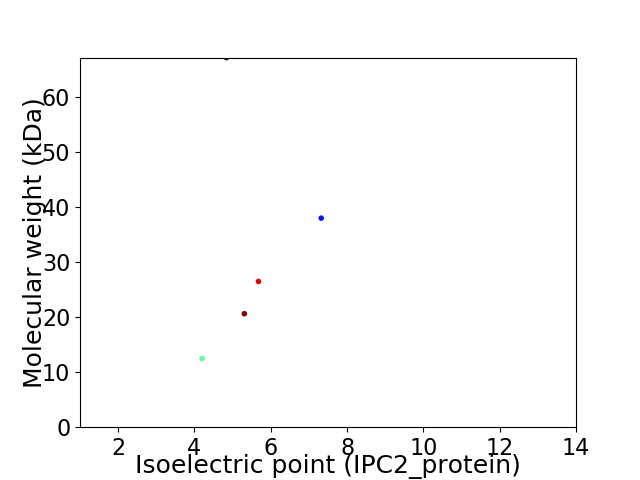
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVQ8|A0A4V1FVQ8_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_188 OX=2585406 PE=3 SV=1
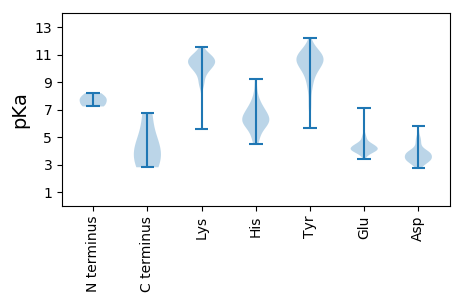
MM1 pKa = 7.79AKK3 pKa = 9.99HH4 pKa = 5.98NYY6 pKa = 9.01YY7 pKa = 10.71SIVLRR12 pKa = 11.84NVQDD16 pKa = 4.17VEE18 pKa = 5.08DD19 pKa = 4.02SDD21 pKa = 5.62SNFFFDD27 pKa = 3.78RR28 pKa = 11.84PFYY31 pKa = 11.3AEE33 pKa = 3.66TDD35 pKa = 3.34AMAKK39 pKa = 10.26LIFEE43 pKa = 5.01ASVCSNPVYY52 pKa = 11.33SMLGDD57 pKa = 5.14DD58 pKa = 3.83IALFKK63 pKa = 10.6IGQFDD68 pKa = 3.95DD69 pKa = 3.93EE70 pKa = 4.59EE71 pKa = 7.08GIMFNIDD78 pKa = 3.12GFDD81 pKa = 3.48EE82 pKa = 4.55LFIMTGSEE90 pKa = 3.78ALNVHH95 pKa = 6.06NEE97 pKa = 3.75RR98 pKa = 11.84SEE100 pKa = 4.42KK101 pKa = 10.23NGKK104 pKa = 8.96DD105 pKa = 3.16GKK107 pKa = 10.16VANN110 pKa = 4.43
MM1 pKa = 7.79AKK3 pKa = 9.99HH4 pKa = 5.98NYY6 pKa = 9.01YY7 pKa = 10.71SIVLRR12 pKa = 11.84NVQDD16 pKa = 4.17VEE18 pKa = 5.08DD19 pKa = 4.02SDD21 pKa = 5.62SNFFFDD27 pKa = 3.78RR28 pKa = 11.84PFYY31 pKa = 11.3AEE33 pKa = 3.66TDD35 pKa = 3.34AMAKK39 pKa = 10.26LIFEE43 pKa = 5.01ASVCSNPVYY52 pKa = 11.33SMLGDD57 pKa = 5.14DD58 pKa = 3.83IALFKK63 pKa = 10.6IGQFDD68 pKa = 3.95DD69 pKa = 3.93EE70 pKa = 4.59EE71 pKa = 7.08GIMFNIDD78 pKa = 3.12GFDD81 pKa = 3.48EE82 pKa = 4.55LFIMTGSEE90 pKa = 3.78ALNVHH95 pKa = 6.06NEE97 pKa = 3.75RR98 pKa = 11.84SEE100 pKa = 4.42KK101 pKa = 10.23NGKK104 pKa = 8.96DD105 pKa = 3.16GKK107 pKa = 10.16VANN110 pKa = 4.43
Molecular weight: 12.47 kDa
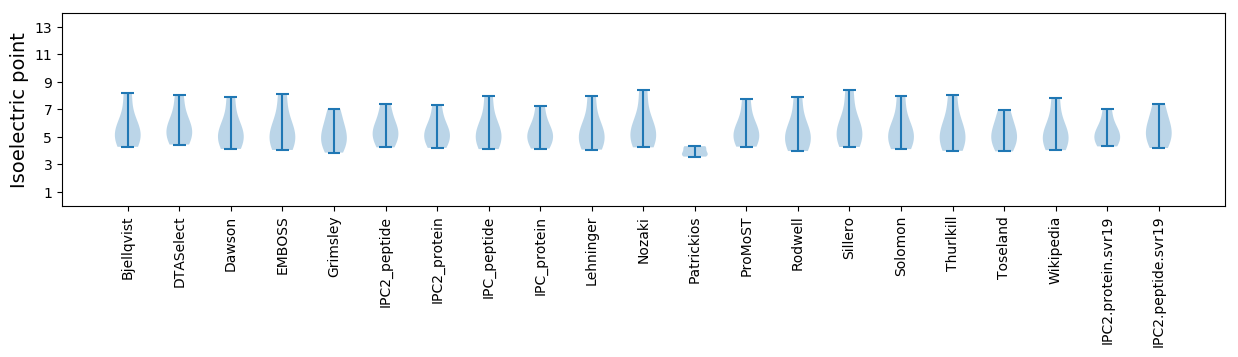
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4H5|A0A4P8W4H5_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_188 OX=2585406 PE=4 SV=1
MM1 pKa = 7.35ARR3 pKa = 11.84WKK5 pKa = 8.87TAEE8 pKa = 4.07GKK10 pKa = 10.29KK11 pKa = 5.6VTRR14 pKa = 11.84YY15 pKa = 9.83IGTSGNIQGIPLDD28 pKa = 3.62AEE30 pKa = 4.44YY31 pKa = 10.74FSVGCGHH38 pKa = 7.61CIACRR43 pKa = 11.84LSKK46 pKa = 11.04SKK48 pKa = 10.76DD49 pKa = 2.78WANRR53 pKa = 11.84MTLEE57 pKa = 3.97AMYY60 pKa = 10.39HH61 pKa = 5.3SSCFFLTLTYY71 pKa = 11.05DD72 pKa = 3.81DD73 pKa = 3.82AHH75 pKa = 8.31LPEE78 pKa = 4.11QHH80 pKa = 5.99LQKK83 pKa = 10.98NGDD86 pKa = 3.2ISPIHH91 pKa = 6.3SLNKK95 pKa = 10.02KK96 pKa = 9.44HH97 pKa = 6.39FQDD100 pKa = 3.38FMKK103 pKa = 10.54RR104 pKa = 11.84LRR106 pKa = 11.84WKK108 pKa = 10.45FRR110 pKa = 11.84DD111 pKa = 3.41VEE113 pKa = 3.84IRR115 pKa = 11.84YY116 pKa = 7.41YY117 pKa = 11.1ACGEE121 pKa = 4.04YY122 pKa = 10.64GEE124 pKa = 4.94KK125 pKa = 10.42SMRR128 pKa = 11.84PHH130 pKa = 4.8YY131 pKa = 10.08HH132 pKa = 6.93AIIFGLPLDD141 pKa = 4.12RR142 pKa = 11.84LGDD145 pKa = 4.52AIYY148 pKa = 10.29SGCSSGFKK156 pKa = 10.07VFAYY160 pKa = 9.77TEE162 pKa = 4.37LSKK165 pKa = 11.09LWVFGNHH172 pKa = 5.44IVAEE176 pKa = 4.48ANWQTMAYY184 pKa = 7.51TSRR187 pKa = 11.84YY188 pKa = 5.63ITKK191 pKa = 9.0KK192 pKa = 10.11QFGIGSKK199 pKa = 9.59IYY201 pKa = 10.43EE202 pKa = 4.16EE203 pKa = 4.28LGYY206 pKa = 10.22IPEE209 pKa = 4.36FSLMSRR215 pKa = 11.84KK216 pKa = 9.59PGIGKK221 pKa = 8.81IWLDD225 pKa = 3.23ASRR228 pKa = 11.84YY229 pKa = 9.75SVLDD233 pKa = 3.71DD234 pKa = 3.52KK235 pKa = 11.52AIYY238 pKa = 10.08IPGKK242 pKa = 9.53GKK244 pKa = 10.16QPIPRR249 pKa = 11.84YY250 pKa = 9.49FKK252 pKa = 10.92EE253 pKa = 3.95KK254 pKa = 10.79LKK256 pKa = 10.54DD257 pKa = 2.88ICYY260 pKa = 9.96FCDD263 pKa = 3.16VDD265 pKa = 5.26GVINPTDD272 pKa = 3.3EE273 pKa = 4.71ALWLEE278 pKa = 4.14FDD280 pKa = 3.4DD281 pKa = 3.9WLRR284 pKa = 11.84DD285 pKa = 3.72VGFHH289 pKa = 6.07QYY291 pKa = 10.72CEE293 pKa = 4.05QQMKK297 pKa = 10.56LDD299 pKa = 3.94QTTLTYY305 pKa = 10.71SEE307 pKa = 4.45VLEE310 pKa = 4.23NTQYY314 pKa = 11.63AHH316 pKa = 6.06EE317 pKa = 4.27QVASSLGRR325 pKa = 11.84KK326 pKa = 8.4GVV328 pKa = 3.16
MM1 pKa = 7.35ARR3 pKa = 11.84WKK5 pKa = 8.87TAEE8 pKa = 4.07GKK10 pKa = 10.29KK11 pKa = 5.6VTRR14 pKa = 11.84YY15 pKa = 9.83IGTSGNIQGIPLDD28 pKa = 3.62AEE30 pKa = 4.44YY31 pKa = 10.74FSVGCGHH38 pKa = 7.61CIACRR43 pKa = 11.84LSKK46 pKa = 11.04SKK48 pKa = 10.76DD49 pKa = 2.78WANRR53 pKa = 11.84MTLEE57 pKa = 3.97AMYY60 pKa = 10.39HH61 pKa = 5.3SSCFFLTLTYY71 pKa = 11.05DD72 pKa = 3.81DD73 pKa = 3.82AHH75 pKa = 8.31LPEE78 pKa = 4.11QHH80 pKa = 5.99LQKK83 pKa = 10.98NGDD86 pKa = 3.2ISPIHH91 pKa = 6.3SLNKK95 pKa = 10.02KK96 pKa = 9.44HH97 pKa = 6.39FQDD100 pKa = 3.38FMKK103 pKa = 10.54RR104 pKa = 11.84LRR106 pKa = 11.84WKK108 pKa = 10.45FRR110 pKa = 11.84DD111 pKa = 3.41VEE113 pKa = 3.84IRR115 pKa = 11.84YY116 pKa = 7.41YY117 pKa = 11.1ACGEE121 pKa = 4.04YY122 pKa = 10.64GEE124 pKa = 4.94KK125 pKa = 10.42SMRR128 pKa = 11.84PHH130 pKa = 4.8YY131 pKa = 10.08HH132 pKa = 6.93AIIFGLPLDD141 pKa = 4.12RR142 pKa = 11.84LGDD145 pKa = 4.52AIYY148 pKa = 10.29SGCSSGFKK156 pKa = 10.07VFAYY160 pKa = 9.77TEE162 pKa = 4.37LSKK165 pKa = 11.09LWVFGNHH172 pKa = 5.44IVAEE176 pKa = 4.48ANWQTMAYY184 pKa = 7.51TSRR187 pKa = 11.84YY188 pKa = 5.63ITKK191 pKa = 9.0KK192 pKa = 10.11QFGIGSKK199 pKa = 9.59IYY201 pKa = 10.43EE202 pKa = 4.16EE203 pKa = 4.28LGYY206 pKa = 10.22IPEE209 pKa = 4.36FSLMSRR215 pKa = 11.84KK216 pKa = 9.59PGIGKK221 pKa = 8.81IWLDD225 pKa = 3.23ASRR228 pKa = 11.84YY229 pKa = 9.75SVLDD233 pKa = 3.71DD234 pKa = 3.52KK235 pKa = 11.52AIYY238 pKa = 10.08IPGKK242 pKa = 9.53GKK244 pKa = 10.16QPIPRR249 pKa = 11.84YY250 pKa = 9.49FKK252 pKa = 10.92EE253 pKa = 3.95KK254 pKa = 10.79LKK256 pKa = 10.54DD257 pKa = 2.88ICYY260 pKa = 9.96FCDD263 pKa = 3.16VDD265 pKa = 5.26GVINPTDD272 pKa = 3.3EE273 pKa = 4.71ALWLEE278 pKa = 4.14FDD280 pKa = 3.4DD281 pKa = 3.9WLRR284 pKa = 11.84DD285 pKa = 3.72VGFHH289 pKa = 6.07QYY291 pKa = 10.72CEE293 pKa = 4.05QQMKK297 pKa = 10.56LDD299 pKa = 3.94QTTLTYY305 pKa = 10.71SEE307 pKa = 4.45VLEE310 pKa = 4.23NTQYY314 pKa = 11.63AHH316 pKa = 6.06EE317 pKa = 4.27QVASSLGRR325 pKa = 11.84KK326 pKa = 8.4GVV328 pKa = 3.16
Molecular weight: 37.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1461 |
110 |
598 |
292.2 |
32.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.297 ± 0.926 | 1.3 ± 0.407 |
7.529 ± 0.607 | 5.476 ± 0.501 |
5.339 ± 0.714 | 7.05 ± 0.521 |
2.19 ± 0.439 | 5.681 ± 0.365 |
5.955 ± 1.456 | 6.776 ± 0.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.22 | 5.133 ± 0.95 |
4.107 ± 1.082 | 3.901 ± 0.409 |
4.312 ± 0.443 | 9.446 ± 2.198 |
5.065 ± 0.519 | 5.749 ± 0.755 |
1.437 ± 0.294 | 4.86 ± 0.401 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
