
Streptomyces sp. RKND-216
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
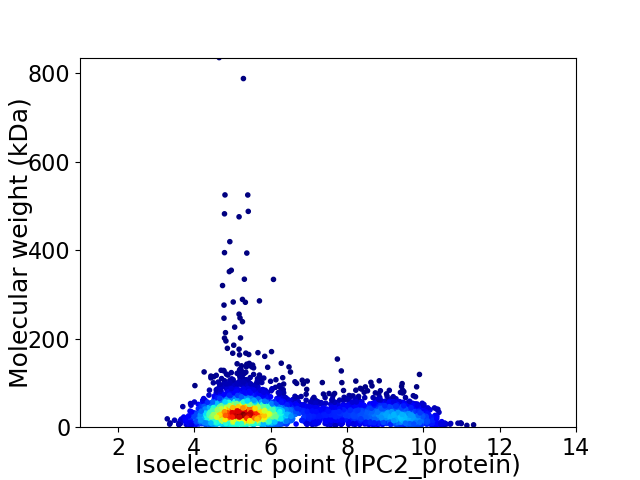
Virtual 2D-PAGE plot for 4607 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S2RFW5|A0A4S2RFW5_9ACTN Foldase YidC OS=Streptomyces sp. RKND-216 OX=2562581 GN=yidC PE=3 SV=1
MM1 pKa = 7.41AAAGALIATALTGPAAGAAPAEE23 pKa = 4.6DD24 pKa = 4.29DD25 pKa = 3.96GSRR28 pKa = 11.84AAAQLLDD35 pKa = 3.62NAGFEE40 pKa = 4.46AGLEE44 pKa = 4.18GWTCSGDD51 pKa = 3.53SGTAAASPARR61 pKa = 11.84SGVRR65 pKa = 11.84ALTATPAGQDD75 pKa = 3.22NARR78 pKa = 11.84CSQVVAVEE86 pKa = 3.99PDD88 pKa = 3.01ADD90 pKa = 3.91YY91 pKa = 10.8TLSGWVQGSYY101 pKa = 10.95VYY103 pKa = 10.78LGATGTGTTDD113 pKa = 2.66VATWTGSTPSWKK125 pKa = 9.81QLSTTFRR132 pKa = 11.84TGPDD136 pKa = 3.27TTSVTVYY143 pKa = 9.75LHH145 pKa = 5.75GWYY148 pKa = 10.23GQGAYY153 pKa = 10.02HH154 pKa = 7.38ADD156 pKa = 3.79DD157 pKa = 4.84LALSGPGEE165 pKa = 4.48PGDD168 pKa = 4.55PGDD171 pKa = 4.49PGDD174 pKa = 4.36PEE176 pKa = 4.48VPAVPSGLAVGAVTAGSVGLSWTEE200 pKa = 3.9VPDD203 pKa = 3.24ATAYY207 pKa = 9.79HH208 pKa = 6.99VYY210 pKa = 10.55RR211 pKa = 11.84DD212 pKa = 3.86GTKK215 pKa = 9.43VASAEE220 pKa = 4.24GASATVTGLSPSTSYY235 pKa = 11.42AFRR238 pKa = 11.84VSAVNGAGEE247 pKa = 4.37SAKK250 pKa = 10.54SAAVTARR257 pKa = 11.84TSADD261 pKa = 3.28GGDD264 pKa = 3.88GGTTVPEE271 pKa = 4.27HH272 pKa = 6.69AVTGYY277 pKa = 9.64VQNFDD282 pKa = 4.67NGAQTQRR289 pKa = 11.84LTEE292 pKa = 3.94VQDD295 pKa = 3.87AYY297 pKa = 11.15DD298 pKa = 4.22IIAVAFASSTSTPGEE313 pKa = 3.93LTFALDD319 pKa = 3.47PALAPSDD326 pKa = 4.08DD327 pKa = 3.91AFKK330 pKa = 11.29ADD332 pKa = 3.33IAAKK336 pKa = 9.28QAEE339 pKa = 4.73GKK341 pKa = 10.38AVILSVGGEE350 pKa = 4.12LGNVTVNDD358 pKa = 4.17DD359 pKa = 3.03ASARR363 pKa = 11.84AFADD367 pKa = 3.69SAWALMQEE375 pKa = 4.24YY376 pKa = 9.84GFDD379 pKa = 4.43GIDD382 pKa = 3.02IDD384 pKa = 4.98LEE386 pKa = 4.23HH387 pKa = 7.34GINSTYY393 pKa = 9.25LTRR396 pKa = 11.84AIRR399 pKa = 11.84DD400 pKa = 3.39LAANAGPDD408 pKa = 3.91FVLTMAPQTLDD419 pKa = 3.33MQSTSTEE426 pKa = 4.27YY427 pKa = 9.12FTLALNVKK435 pKa = 10.3DD436 pKa = 3.51ILTVVNMQYY445 pKa = 11.07YY446 pKa = 10.53NSGSMLGCDD455 pKa = 3.33GQVYY459 pKa = 10.08SQGSVDD465 pKa = 5.27FLTALACIQLRR476 pKa = 11.84GGLDD480 pKa = 3.27ASQVGLGTPASPRR493 pKa = 11.84GAGSGYY499 pKa = 10.36VDD501 pKa = 3.29PSVVNDD507 pKa = 4.17ALSCLTRR514 pKa = 11.84GTNCGSFQPSQTYY527 pKa = 9.41PSLRR531 pKa = 11.84GAMTWSTNWDD541 pKa = 3.96ALNGDD546 pKa = 3.36QWSNAVGPHH555 pKa = 4.7VHH557 pKa = 6.75GLPP560 pKa = 3.71
MM1 pKa = 7.41AAAGALIATALTGPAAGAAPAEE23 pKa = 4.6DD24 pKa = 4.29DD25 pKa = 3.96GSRR28 pKa = 11.84AAAQLLDD35 pKa = 3.62NAGFEE40 pKa = 4.46AGLEE44 pKa = 4.18GWTCSGDD51 pKa = 3.53SGTAAASPARR61 pKa = 11.84SGVRR65 pKa = 11.84ALTATPAGQDD75 pKa = 3.22NARR78 pKa = 11.84CSQVVAVEE86 pKa = 3.99PDD88 pKa = 3.01ADD90 pKa = 3.91YY91 pKa = 10.8TLSGWVQGSYY101 pKa = 10.95VYY103 pKa = 10.78LGATGTGTTDD113 pKa = 2.66VATWTGSTPSWKK125 pKa = 9.81QLSTTFRR132 pKa = 11.84TGPDD136 pKa = 3.27TTSVTVYY143 pKa = 9.75LHH145 pKa = 5.75GWYY148 pKa = 10.23GQGAYY153 pKa = 10.02HH154 pKa = 7.38ADD156 pKa = 3.79DD157 pKa = 4.84LALSGPGEE165 pKa = 4.48PGDD168 pKa = 4.55PGDD171 pKa = 4.49PGDD174 pKa = 4.36PEE176 pKa = 4.48VPAVPSGLAVGAVTAGSVGLSWTEE200 pKa = 3.9VPDD203 pKa = 3.24ATAYY207 pKa = 9.79HH208 pKa = 6.99VYY210 pKa = 10.55RR211 pKa = 11.84DD212 pKa = 3.86GTKK215 pKa = 9.43VASAEE220 pKa = 4.24GASATVTGLSPSTSYY235 pKa = 11.42AFRR238 pKa = 11.84VSAVNGAGEE247 pKa = 4.37SAKK250 pKa = 10.54SAAVTARR257 pKa = 11.84TSADD261 pKa = 3.28GGDD264 pKa = 3.88GGTTVPEE271 pKa = 4.27HH272 pKa = 6.69AVTGYY277 pKa = 9.64VQNFDD282 pKa = 4.67NGAQTQRR289 pKa = 11.84LTEE292 pKa = 3.94VQDD295 pKa = 3.87AYY297 pKa = 11.15DD298 pKa = 4.22IIAVAFASSTSTPGEE313 pKa = 3.93LTFALDD319 pKa = 3.47PALAPSDD326 pKa = 4.08DD327 pKa = 3.91AFKK330 pKa = 11.29ADD332 pKa = 3.33IAAKK336 pKa = 9.28QAEE339 pKa = 4.73GKK341 pKa = 10.38AVILSVGGEE350 pKa = 4.12LGNVTVNDD358 pKa = 4.17DD359 pKa = 3.03ASARR363 pKa = 11.84AFADD367 pKa = 3.69SAWALMQEE375 pKa = 4.24YY376 pKa = 9.84GFDD379 pKa = 4.43GIDD382 pKa = 3.02IDD384 pKa = 4.98LEE386 pKa = 4.23HH387 pKa = 7.34GINSTYY393 pKa = 9.25LTRR396 pKa = 11.84AIRR399 pKa = 11.84DD400 pKa = 3.39LAANAGPDD408 pKa = 3.91FVLTMAPQTLDD419 pKa = 3.33MQSTSTEE426 pKa = 4.27YY427 pKa = 9.12FTLALNVKK435 pKa = 10.3DD436 pKa = 3.51ILTVVNMQYY445 pKa = 11.07YY446 pKa = 10.53NSGSMLGCDD455 pKa = 3.33GQVYY459 pKa = 10.08SQGSVDD465 pKa = 5.27FLTALACIQLRR476 pKa = 11.84GGLDD480 pKa = 3.27ASQVGLGTPASPRR493 pKa = 11.84GAGSGYY499 pKa = 10.36VDD501 pKa = 3.29PSVVNDD507 pKa = 4.17ALSCLTRR514 pKa = 11.84GTNCGSFQPSQTYY527 pKa = 9.41PSLRR531 pKa = 11.84GAMTWSTNWDD541 pKa = 3.96ALNGDD546 pKa = 3.36QWSNAVGPHH555 pKa = 4.7VHH557 pKa = 6.75GLPP560 pKa = 3.71
Molecular weight: 56.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S2R4B0|A0A4S2R4B0_9ACTN CDP-alcohol phosphatidyltransferase family protein OS=Streptomyces sp. RKND-216 OX=2562581 GN=E4198_00700 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.81GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.81GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1585695 |
25 |
8058 |
344.2 |
36.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.803 ± 0.065 | 0.827 ± 0.01 |
6.214 ± 0.034 | 6.009 ± 0.035 |
2.666 ± 0.02 | 9.862 ± 0.046 |
2.418 ± 0.021 | 2.663 ± 0.025 |
1.843 ± 0.027 | 10.287 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.706 ± 0.016 | 1.57 ± 0.016 |
6.281 ± 0.034 | 2.638 ± 0.021 |
8.53 ± 0.046 | 4.819 ± 0.026 |
5.93 ± 0.039 | 8.539 ± 0.037 |
1.441 ± 0.02 | 1.953 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
