
Flavobacterium humi
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
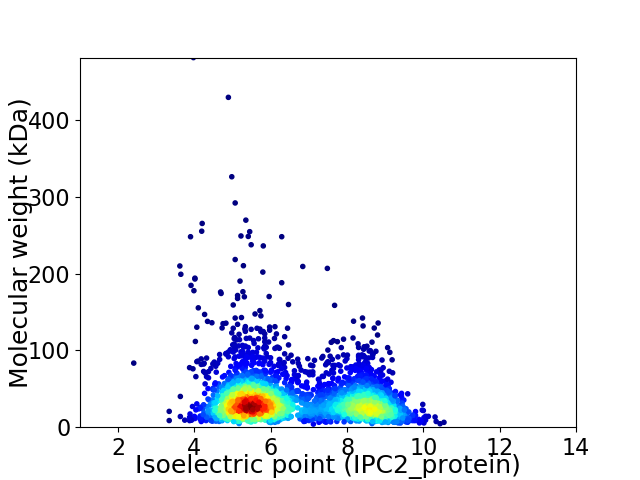
Virtual 2D-PAGE plot for 3054 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z0L6N4|A0A4Z0L6N4_9FLAO Uncharacterized protein OS=Flavobacterium humi OX=2562683 GN=E4635_15100 PE=4 SV=1
MM1 pKa = 7.25NKK3 pKa = 9.97KK4 pKa = 10.33LLFLFIFFFLGLASVVSQNVSLYY27 pKa = 9.23SQFNGRR33 pKa = 11.84YY34 pKa = 9.07DD35 pKa = 3.35FAFIGNTLNTNEE47 pKa = 4.63NGTGAPCTITTSSSATVNLNPSDD70 pKa = 3.96MVEE73 pKa = 3.57AAYY76 pKa = 10.1LYY78 pKa = 9.41WAGSGTGDD86 pKa = 4.7FDD88 pKa = 4.65VKK90 pKa = 11.13LNDD93 pKa = 3.41VAITATRR100 pKa = 11.84QFSVIQPTSQRR111 pKa = 11.84PFFSAFADD119 pKa = 3.04ITTQVQTSGNGNYY132 pKa = 8.8TLSEE136 pKa = 4.25LDD138 pKa = 3.62LTNVIAQYY146 pKa = 10.65CPNGTNFGGWAIIIVYY162 pKa = 10.08KK163 pKa = 10.78NAALPLNQLNIYY175 pKa = 10.35DD176 pKa = 4.24GLQFVPDD183 pKa = 5.54AINITLDD190 pKa = 3.4SLNVIDD196 pKa = 6.22NEE198 pKa = 4.08DD199 pKa = 3.32AKK201 pKa = 10.86IGFVAWEE208 pKa = 4.23GDD210 pKa = 3.1RR211 pKa = 11.84SIAEE215 pKa = 4.1NEE217 pKa = 4.08SLRR220 pKa = 11.84INGNLISNPPLNPSDD235 pKa = 3.48NAFNGTNSFTGSSVLYY251 pKa = 11.22NMDD254 pKa = 4.0LDD256 pKa = 4.39VYY258 pKa = 10.73NIQNNIQIGDD268 pKa = 3.76EE269 pKa = 4.15SASIQLTSDD278 pKa = 3.18RR279 pKa = 11.84DD280 pKa = 3.64FVMINAIVTKK290 pKa = 10.5LNSQLPDD297 pKa = 2.95ATVAIEE303 pKa = 5.66NIAQEE308 pKa = 4.53CNTRR312 pKa = 11.84RR313 pKa = 11.84LTIQYY318 pKa = 10.17SIRR321 pKa = 11.84NFNATDD327 pKa = 4.11FLPSGTFTTIYY338 pKa = 10.69ADD340 pKa = 4.94GIAVGVVQTTEE351 pKa = 4.88DD352 pKa = 3.35IPIGEE357 pKa = 4.23TRR359 pKa = 11.84NYY361 pKa = 6.72EE362 pKa = 4.01TTVIIPASVAPVFDD376 pKa = 3.78LQFIVDD382 pKa = 3.96NEE384 pKa = 4.17NGVAHH389 pKa = 4.88VTEE392 pKa = 4.51TNEE395 pKa = 3.87NNNTFTQEE403 pKa = 3.64VALWLSPEE411 pKa = 3.67INAPQNVTACNEE423 pKa = 3.86GNTRR427 pKa = 11.84GTFDD431 pKa = 4.92FSLQTGAIPINPGDD445 pKa = 3.39TVLFFEE451 pKa = 4.74NNQDD455 pKa = 3.47AVLSQNPIINTTNYY469 pKa = 9.74LAQATPKK476 pKa = 9.12TIYY479 pKa = 10.38VRR481 pKa = 11.84IEE483 pKa = 3.75NEE485 pKa = 3.64HH486 pKa = 6.84CFTITHH492 pKa = 6.61FEE494 pKa = 4.13LVAIPCPPTVYY505 pKa = 10.83NYY507 pKa = 7.52FTPNGDD513 pKa = 3.62GLNDD517 pKa = 3.59SFFVEE522 pKa = 4.58GLRR525 pKa = 11.84DD526 pKa = 3.64VFLHH530 pKa = 6.54FEE532 pKa = 3.91LSIYY536 pKa = 9.76NRR538 pKa = 11.84WGSLVWEE545 pKa = 4.84GNNNTEE551 pKa = 4.23DD552 pKa = 3.39WDD554 pKa = 4.21GKK556 pKa = 10.39SNQPAILFHH565 pKa = 6.79GNLPEE570 pKa = 3.96GTYY573 pKa = 10.27FYY575 pKa = 11.43VLYY578 pKa = 10.17LNEE581 pKa = 4.78PGYY584 pKa = 9.89PSPLTGWVFMNRR596 pKa = 3.4
MM1 pKa = 7.25NKK3 pKa = 9.97KK4 pKa = 10.33LLFLFIFFFLGLASVVSQNVSLYY27 pKa = 9.23SQFNGRR33 pKa = 11.84YY34 pKa = 9.07DD35 pKa = 3.35FAFIGNTLNTNEE47 pKa = 4.63NGTGAPCTITTSSSATVNLNPSDD70 pKa = 3.96MVEE73 pKa = 3.57AAYY76 pKa = 10.1LYY78 pKa = 9.41WAGSGTGDD86 pKa = 4.7FDD88 pKa = 4.65VKK90 pKa = 11.13LNDD93 pKa = 3.41VAITATRR100 pKa = 11.84QFSVIQPTSQRR111 pKa = 11.84PFFSAFADD119 pKa = 3.04ITTQVQTSGNGNYY132 pKa = 8.8TLSEE136 pKa = 4.25LDD138 pKa = 3.62LTNVIAQYY146 pKa = 10.65CPNGTNFGGWAIIIVYY162 pKa = 10.08KK163 pKa = 10.78NAALPLNQLNIYY175 pKa = 10.35DD176 pKa = 4.24GLQFVPDD183 pKa = 5.54AINITLDD190 pKa = 3.4SLNVIDD196 pKa = 6.22NEE198 pKa = 4.08DD199 pKa = 3.32AKK201 pKa = 10.86IGFVAWEE208 pKa = 4.23GDD210 pKa = 3.1RR211 pKa = 11.84SIAEE215 pKa = 4.1NEE217 pKa = 4.08SLRR220 pKa = 11.84INGNLISNPPLNPSDD235 pKa = 3.48NAFNGTNSFTGSSVLYY251 pKa = 11.22NMDD254 pKa = 4.0LDD256 pKa = 4.39VYY258 pKa = 10.73NIQNNIQIGDD268 pKa = 3.76EE269 pKa = 4.15SASIQLTSDD278 pKa = 3.18RR279 pKa = 11.84DD280 pKa = 3.64FVMINAIVTKK290 pKa = 10.5LNSQLPDD297 pKa = 2.95ATVAIEE303 pKa = 5.66NIAQEE308 pKa = 4.53CNTRR312 pKa = 11.84RR313 pKa = 11.84LTIQYY318 pKa = 10.17SIRR321 pKa = 11.84NFNATDD327 pKa = 4.11FLPSGTFTTIYY338 pKa = 10.69ADD340 pKa = 4.94GIAVGVVQTTEE351 pKa = 4.88DD352 pKa = 3.35IPIGEE357 pKa = 4.23TRR359 pKa = 11.84NYY361 pKa = 6.72EE362 pKa = 4.01TTVIIPASVAPVFDD376 pKa = 3.78LQFIVDD382 pKa = 3.96NEE384 pKa = 4.17NGVAHH389 pKa = 4.88VTEE392 pKa = 4.51TNEE395 pKa = 3.87NNNTFTQEE403 pKa = 3.64VALWLSPEE411 pKa = 3.67INAPQNVTACNEE423 pKa = 3.86GNTRR427 pKa = 11.84GTFDD431 pKa = 4.92FSLQTGAIPINPGDD445 pKa = 3.39TVLFFEE451 pKa = 4.74NNQDD455 pKa = 3.47AVLSQNPIINTTNYY469 pKa = 9.74LAQATPKK476 pKa = 9.12TIYY479 pKa = 10.38VRR481 pKa = 11.84IEE483 pKa = 3.75NEE485 pKa = 3.64HH486 pKa = 6.84CFTITHH492 pKa = 6.61FEE494 pKa = 4.13LVAIPCPPTVYY505 pKa = 10.83NYY507 pKa = 7.52FTPNGDD513 pKa = 3.62GLNDD517 pKa = 3.59SFFVEE522 pKa = 4.58GLRR525 pKa = 11.84DD526 pKa = 3.64VFLHH530 pKa = 6.54FEE532 pKa = 3.91LSIYY536 pKa = 9.76NRR538 pKa = 11.84WGSLVWEE545 pKa = 4.84GNNNTEE551 pKa = 4.23DD552 pKa = 3.39WDD554 pKa = 4.21GKK556 pKa = 10.39SNQPAILFHH565 pKa = 6.79GNLPEE570 pKa = 3.96GTYY573 pKa = 10.27FYY575 pKa = 11.43VLYY578 pKa = 10.17LNEE581 pKa = 4.78PGYY584 pKa = 9.89PSPLTGWVFMNRR596 pKa = 3.4
Molecular weight: 65.82 kDa
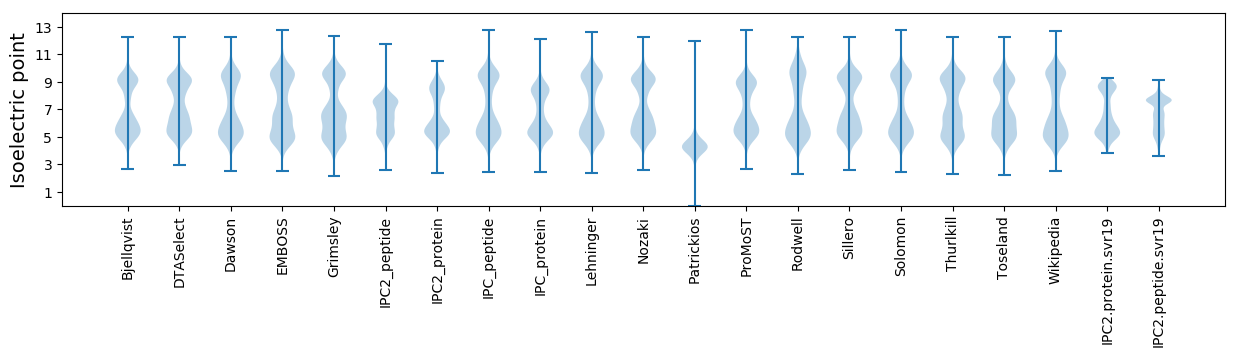
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z0L5Y1|A0A4Z0L5Y1_9FLAO Phage tail protein OS=Flavobacterium humi OX=2562683 GN=E4635_11020 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.09KK28 pKa = 11.02PEE30 pKa = 4.34RR31 pKa = 11.84SLIAPIKK38 pKa = 10.47NSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.86KK59 pKa = 9.44QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.24RR69 pKa = 11.84TKK71 pKa = 10.49AGIPATVKK79 pKa = 10.65SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLAYY96 pKa = 10.31ADD98 pKa = 3.96GEE100 pKa = 4.24KK101 pKa = 10.14TYY103 pKa = 10.54IIAQNGLQVGQKK115 pKa = 9.1VVSGEE120 pKa = 3.88GSAPEE125 pKa = 4.11IGNTLPLSKK134 pKa = 10.28IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCSATIGAVSNSDD199 pKa = 2.96HH200 pKa = 6.02QLIVSGKK207 pKa = 9.19AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.81PRR245 pKa = 11.84SRR247 pKa = 11.84KK248 pKa = 8.85GLPAKK253 pKa = 10.34GYY255 pKa = 7.39RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.74QNPSNKK266 pKa = 10.03YY267 pKa = 8.27IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.09KK28 pKa = 11.02PEE30 pKa = 4.34RR31 pKa = 11.84SLIAPIKK38 pKa = 10.47NSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.86KK59 pKa = 9.44QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.24RR69 pKa = 11.84TKK71 pKa = 10.49AGIPATVKK79 pKa = 10.65SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLAYY96 pKa = 10.31ADD98 pKa = 3.96GEE100 pKa = 4.24KK101 pKa = 10.14TYY103 pKa = 10.54IIAQNGLQVGQKK115 pKa = 9.1VVSGEE120 pKa = 3.88GSAPEE125 pKa = 4.11IGNTLPLSKK134 pKa = 10.28IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCSATIGAVSNSDD199 pKa = 2.96HH200 pKa = 6.02QLIVSGKK207 pKa = 9.19AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.81PRR245 pKa = 11.84SRR247 pKa = 11.84KK248 pKa = 8.85GLPAKK253 pKa = 10.34GYY255 pKa = 7.39RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.74QNPSNKK266 pKa = 10.03YY267 pKa = 8.27IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
Molecular weight: 29.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1069261 |
33 |
4662 |
350.1 |
39.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.887 ± 0.05 | 0.843 ± 0.019 |
5.265 ± 0.033 | 6.314 ± 0.058 |
5.284 ± 0.04 | 6.503 ± 0.049 |
1.79 ± 0.025 | 7.658 ± 0.038 |
7.656 ± 0.069 | 9.001 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.211 ± 0.026 | 6.141 ± 0.05 |
3.589 ± 0.031 | 3.586 ± 0.027 |
3.227 ± 0.03 | 6.497 ± 0.04 |
6.199 ± 0.078 | 6.26 ± 0.038 |
1.008 ± 0.014 | 4.083 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
