
Lumpfish ranavirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Iridoviridae; Alphairidovirinae; Ranavirus; European North Atlantic ranavirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
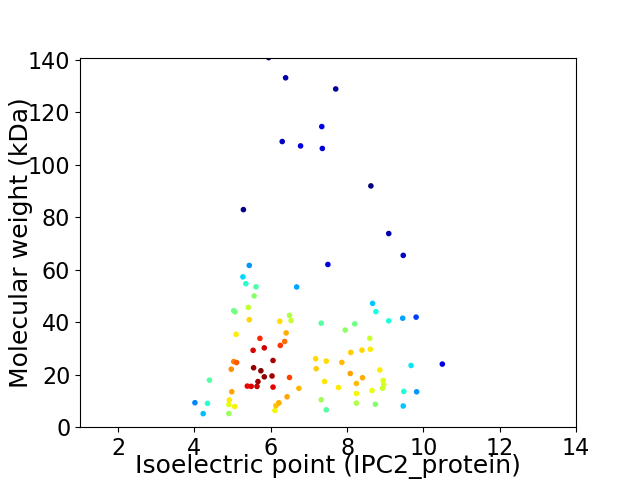
Virtual 2D-PAGE plot for 97 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9T8Q8|A0A3Q9T8Q8_9VIRU NTPase OS=Lumpfish ranavirus OX=2501771 GN=ORF8 PE=4 SV=1
MM1 pKa = 7.83ANTQNSVMDD10 pKa = 4.01FVDD13 pKa = 3.5TNVIEE18 pKa = 4.34VVRR21 pKa = 11.84DD22 pKa = 3.31QGGLYY27 pKa = 10.28HH28 pKa = 7.1VILEE32 pKa = 4.57KK33 pKa = 10.36ATEE36 pKa = 3.83LAIPDD41 pKa = 3.93EE42 pKa = 4.81YY43 pKa = 10.73ILEE46 pKa = 4.37DD47 pKa = 4.89FYY49 pKa = 11.82SDD51 pKa = 3.69TYY53 pKa = 9.96KK54 pKa = 10.6HH55 pKa = 5.21QVMVLEE61 pKa = 5.29KK62 pKa = 10.22IPKK65 pKa = 8.94VWADD69 pKa = 3.0DD70 pKa = 3.16WGYY73 pKa = 11.33DD74 pKa = 3.65EE75 pKa = 6.0NVLEE79 pKa = 4.37EE80 pKa = 4.16
MM1 pKa = 7.83ANTQNSVMDD10 pKa = 4.01FVDD13 pKa = 3.5TNVIEE18 pKa = 4.34VVRR21 pKa = 11.84DD22 pKa = 3.31QGGLYY27 pKa = 10.28HH28 pKa = 7.1VILEE32 pKa = 4.57KK33 pKa = 10.36ATEE36 pKa = 3.83LAIPDD41 pKa = 3.93EE42 pKa = 4.81YY43 pKa = 10.73ILEE46 pKa = 4.37DD47 pKa = 4.89FYY49 pKa = 11.82SDD51 pKa = 3.69TYY53 pKa = 9.96KK54 pKa = 10.6HH55 pKa = 5.21QVMVLEE61 pKa = 5.29KK62 pKa = 10.22IPKK65 pKa = 8.94VWADD69 pKa = 3.0DD70 pKa = 3.16WGYY73 pKa = 11.33DD74 pKa = 3.65EE75 pKa = 6.0NVLEE79 pKa = 4.37EE80 pKa = 4.16
Molecular weight: 9.34 kDa
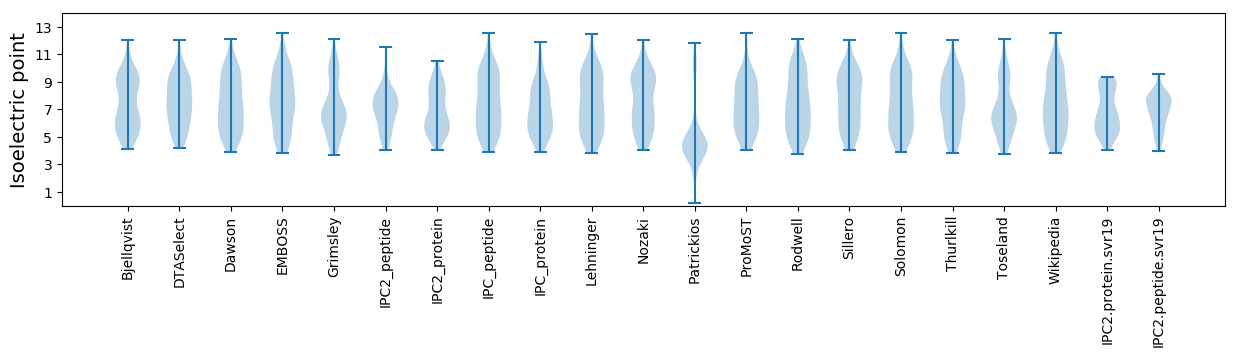
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9T7F8|A0A3Q9T7F8_9VIRU Uncharacterized protein OS=Lumpfish ranavirus OX=2501771 GN=ORF36 PE=4 SV=1
MM1 pKa = 7.64LLHH4 pKa = 7.26LILNLKK10 pKa = 10.3SFLPKK15 pKa = 9.98RR16 pKa = 11.84NLAFRR21 pKa = 11.84YY22 pKa = 10.25DD23 pKa = 3.59IVALSLTHH31 pKa = 7.37KK32 pKa = 10.31ILLKK36 pKa = 10.31HH37 pKa = 5.64QPPEE41 pKa = 3.88NVFAALYY48 pKa = 9.96IDD50 pKa = 4.0SLHH53 pKa = 6.61GGRR56 pKa = 11.84HH57 pKa = 4.65QIMPGFGKK65 pKa = 10.51LSCPTLDD72 pKa = 3.31VGPGLGRR79 pKa = 11.84FAASHH84 pKa = 5.59FSEE87 pKa = 5.14RR88 pKa = 11.84VSQDD92 pKa = 2.44RR93 pKa = 11.84QAQGRR98 pKa = 11.84EE99 pKa = 3.81RR100 pKa = 11.84RR101 pKa = 11.84SVLLSQEE108 pKa = 3.96RR109 pKa = 11.84RR110 pKa = 11.84GSSGRR115 pKa = 11.84QPLYY119 pKa = 10.27SLLPHH124 pKa = 6.15RR125 pKa = 11.84PKK127 pKa = 10.66RR128 pKa = 11.84EE129 pKa = 3.88GVIGAALCYY138 pKa = 9.71TSASHH143 pKa = 6.32SLSPAALLPPVGRR156 pKa = 11.84GKK158 pKa = 10.22DD159 pKa = 3.36RR160 pKa = 11.84HH161 pKa = 6.76CGRR164 pKa = 11.84QVGAAGLLRR173 pKa = 11.84HH174 pKa = 5.84VGRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84QFAQLTRR186 pKa = 11.84GPLGLPCALLEE197 pKa = 4.35PPPLDD202 pKa = 3.32PHH204 pKa = 7.83LLDD207 pKa = 3.87KK208 pKa = 11.36GRR210 pKa = 11.84DD211 pKa = 3.52VGVGGCHH218 pKa = 6.38DD219 pKa = 3.93HH220 pKa = 7.14QGQEE224 pKa = 4.61DD225 pKa = 4.34GLLLHH230 pKa = 6.99GGPEE234 pKa = 4.27PVLGPQGVIGPAYY247 pKa = 9.79GRR249 pKa = 11.84PHH251 pKa = 7.16SNVHH255 pKa = 5.19ALGYY259 pKa = 9.26EE260 pKa = 4.22LQHH263 pKa = 6.64GPLGDD268 pKa = 3.84PPRR271 pKa = 11.84LRR273 pKa = 11.84SGRR276 pKa = 11.84HH277 pKa = 4.81LRR279 pKa = 11.84TFGIVFQRR287 pKa = 11.84VFQRR291 pKa = 11.84VFQRR295 pKa = 11.84VSRR298 pKa = 11.84RR299 pKa = 11.84RR300 pKa = 11.84FGGCFGGTSAVPRR313 pKa = 11.84RR314 pKa = 11.84NVVQEE319 pKa = 3.96SFNVHH324 pKa = 6.78DD325 pKa = 4.76EE326 pKa = 4.3SEE328 pKa = 4.06KK329 pKa = 10.86DD330 pKa = 3.54SRR332 pKa = 11.84GMCPSRR338 pKa = 11.84TVRR341 pKa = 11.84PFQKK345 pKa = 10.14VSHH348 pKa = 6.59LSGGEE353 pKa = 3.5RR354 pKa = 11.84WEE356 pKa = 4.06RR357 pKa = 11.84NYY359 pKa = 10.78PLLFCTLCDD368 pKa = 3.56QLEE371 pKa = 4.23LLGVQQ376 pKa = 3.94
MM1 pKa = 7.64LLHH4 pKa = 7.26LILNLKK10 pKa = 10.3SFLPKK15 pKa = 9.98RR16 pKa = 11.84NLAFRR21 pKa = 11.84YY22 pKa = 10.25DD23 pKa = 3.59IVALSLTHH31 pKa = 7.37KK32 pKa = 10.31ILLKK36 pKa = 10.31HH37 pKa = 5.64QPPEE41 pKa = 3.88NVFAALYY48 pKa = 9.96IDD50 pKa = 4.0SLHH53 pKa = 6.61GGRR56 pKa = 11.84HH57 pKa = 4.65QIMPGFGKK65 pKa = 10.51LSCPTLDD72 pKa = 3.31VGPGLGRR79 pKa = 11.84FAASHH84 pKa = 5.59FSEE87 pKa = 5.14RR88 pKa = 11.84VSQDD92 pKa = 2.44RR93 pKa = 11.84QAQGRR98 pKa = 11.84EE99 pKa = 3.81RR100 pKa = 11.84RR101 pKa = 11.84SVLLSQEE108 pKa = 3.96RR109 pKa = 11.84RR110 pKa = 11.84GSSGRR115 pKa = 11.84QPLYY119 pKa = 10.27SLLPHH124 pKa = 6.15RR125 pKa = 11.84PKK127 pKa = 10.66RR128 pKa = 11.84EE129 pKa = 3.88GVIGAALCYY138 pKa = 9.71TSASHH143 pKa = 6.32SLSPAALLPPVGRR156 pKa = 11.84GKK158 pKa = 10.22DD159 pKa = 3.36RR160 pKa = 11.84HH161 pKa = 6.76CGRR164 pKa = 11.84QVGAAGLLRR173 pKa = 11.84HH174 pKa = 5.84VGRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84QFAQLTRR186 pKa = 11.84GPLGLPCALLEE197 pKa = 4.35PPPLDD202 pKa = 3.32PHH204 pKa = 7.83LLDD207 pKa = 3.87KK208 pKa = 11.36GRR210 pKa = 11.84DD211 pKa = 3.52VGVGGCHH218 pKa = 6.38DD219 pKa = 3.93HH220 pKa = 7.14QGQEE224 pKa = 4.61DD225 pKa = 4.34GLLLHH230 pKa = 6.99GGPEE234 pKa = 4.27PVLGPQGVIGPAYY247 pKa = 9.79GRR249 pKa = 11.84PHH251 pKa = 7.16SNVHH255 pKa = 5.19ALGYY259 pKa = 9.26EE260 pKa = 4.22LQHH263 pKa = 6.64GPLGDD268 pKa = 3.84PPRR271 pKa = 11.84LRR273 pKa = 11.84SGRR276 pKa = 11.84HH277 pKa = 4.81LRR279 pKa = 11.84TFGIVFQRR287 pKa = 11.84VFQRR291 pKa = 11.84VFQRR295 pKa = 11.84VSRR298 pKa = 11.84RR299 pKa = 11.84RR300 pKa = 11.84FGGCFGGTSAVPRR313 pKa = 11.84RR314 pKa = 11.84NVVQEE319 pKa = 3.96SFNVHH324 pKa = 6.78DD325 pKa = 4.76EE326 pKa = 4.3SEE328 pKa = 4.06KK329 pKa = 10.86DD330 pKa = 3.54SRR332 pKa = 11.84GMCPSRR338 pKa = 11.84TVRR341 pKa = 11.84PFQKK345 pKa = 10.14VSHH348 pKa = 6.59LSGGEE353 pKa = 3.5RR354 pKa = 11.84WEE356 pKa = 4.06RR357 pKa = 11.84NYY359 pKa = 10.78PLLFCTLCDD368 pKa = 3.56QLEE371 pKa = 4.23LLGVQQ376 pKa = 3.94
Molecular weight: 41.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
30409 |
49 |
1293 |
313.5 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.586 ± 0.227 | 2.013 ± 0.129 |
6.123 ± 0.168 | 5.748 ± 0.18 |
3.265 ± 0.14 | 7.764 ± 0.379 |
2.161 ± 0.146 | 3.242 ± 0.135 |
5.462 ± 0.234 | 7.929 ± 0.274 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.068 ± 0.134 | 2.568 ± 0.13 |
6.242 ± 0.304 | 2.933 ± 0.243 |
7.001 ± 0.265 | 7.159 ± 0.205 |
5.725 ± 0.164 | 8.603 ± 0.225 |
1.388 ± 0.095 | 3.019 ± 0.147 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
