
Avon-Heathcote Estuary associated circular virus 15
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
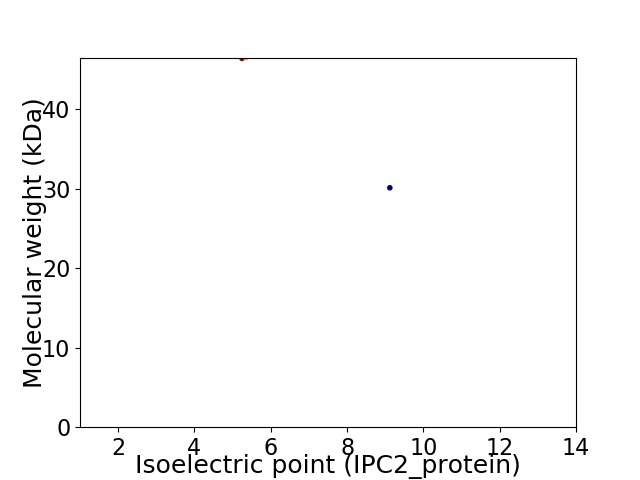
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IB41|A0A0C5IB41_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 15 OX=1618238 PE=3 SV=1
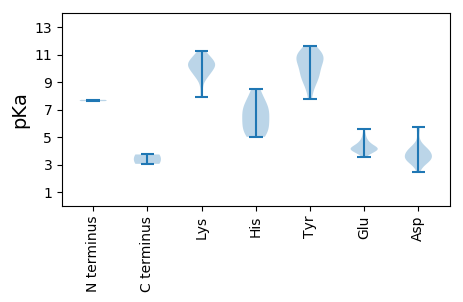
MM1 pKa = 7.61HH2 pKa = 7.1NTTALQSSYY11 pKa = 10.8PNRR14 pKa = 11.84NSRR17 pKa = 11.84SPTANRR23 pKa = 11.84DD24 pKa = 3.09GAIRR28 pKa = 11.84NQSPRR33 pKa = 11.84TQQRR37 pKa = 11.84LSRR40 pKa = 11.84FVFTLNNWTEE50 pKa = 3.8AEE52 pKa = 4.21YY53 pKa = 10.93TDD55 pKa = 5.74IIASDD60 pKa = 4.0VKK62 pKa = 9.59WLVVGKK68 pKa = 10.15EE69 pKa = 3.88IGDD72 pKa = 3.59EE73 pKa = 4.32GTPHH77 pKa = 6.39LQGAVVIGKK86 pKa = 9.26QIAFSTVKK94 pKa = 9.82KK95 pKa = 9.71WPGFSRR101 pKa = 11.84AHH103 pKa = 5.99IEE105 pKa = 3.95NMRR108 pKa = 11.84GKK110 pKa = 9.71PFDD113 pKa = 3.55SLKK116 pKa = 10.9YY117 pKa = 9.23CTKK120 pKa = 10.17QDD122 pKa = 2.72TDD124 pKa = 3.85AFCKK128 pKa = 10.89GEE130 pKa = 4.0MPQEE134 pKa = 4.21GKK136 pKa = 10.82RR137 pKa = 11.84NDD139 pKa = 3.18LHH141 pKa = 7.36DD142 pKa = 5.73AINALKK148 pKa = 10.56QGGTLTSIVKK158 pKa = 10.36SDD160 pKa = 3.96DD161 pKa = 3.27VGLAAAVARR170 pKa = 11.84YY171 pKa = 9.68SKK173 pKa = 10.37GLAMVSSMLKK183 pKa = 9.68PRR185 pKa = 11.84RR186 pKa = 11.84SEE188 pKa = 3.84PPLVIWLSGEE198 pKa = 4.0TGVGKK203 pKa = 8.49TRR205 pKa = 11.84SAVALADD212 pKa = 5.22RR213 pKa = 11.84IAPDD217 pKa = 4.04NYY219 pKa = 9.27WISAGSLRR227 pKa = 11.84WFDD230 pKa = 4.57GYY232 pKa = 10.89DD233 pKa = 3.14GHH235 pKa = 8.5AIAIFDD241 pKa = 4.41DD242 pKa = 4.2LRR244 pKa = 11.84TKK246 pKa = 10.37HH247 pKa = 6.59AEE249 pKa = 3.69FSFLLRR255 pKa = 11.84LLDD258 pKa = 4.3RR259 pKa = 11.84YY260 pKa = 9.37PLRR263 pKa = 11.84VEE265 pKa = 4.48FKK267 pKa = 10.9GGFIDD272 pKa = 3.53WTPSVIVVTAPKK284 pKa = 10.33SPTHH288 pKa = 5.26MWSLRR293 pKa = 11.84NDD295 pKa = 3.27EE296 pKa = 5.6DD297 pKa = 3.61KK298 pKa = 11.24QQLEE302 pKa = 4.26RR303 pKa = 11.84RR304 pKa = 11.84IHH306 pKa = 5.73LAYY309 pKa = 10.36DD310 pKa = 3.44AEE312 pKa = 4.81EE313 pKa = 5.1GDD315 pKa = 3.94DD316 pKa = 4.22WEE318 pKa = 4.78LLLAKK323 pKa = 10.25SEE325 pKa = 4.59AIVAEE330 pKa = 4.54RR331 pKa = 11.84WPTGSYY337 pKa = 8.75PWKK340 pKa = 10.5HH341 pKa = 5.4YY342 pKa = 11.24DD343 pKa = 3.49EE344 pKa = 5.27VNVGSDD350 pKa = 2.93EE351 pKa = 4.17TTIPEE356 pKa = 4.19EE357 pKa = 4.36RR358 pKa = 11.84SCSLSTMGQEE368 pKa = 4.66PDD370 pKa = 3.32TSPSIIEE377 pKa = 4.15IEE379 pKa = 4.05SDD381 pKa = 3.35LSSEE385 pKa = 4.14EE386 pKa = 4.16LVISSSEE393 pKa = 4.07EE394 pKa = 3.53EE395 pKa = 4.18HH396 pKa = 7.74SYY398 pKa = 8.57EE399 pKa = 4.04TQRR402 pKa = 11.84EE403 pKa = 4.34LFSTQYY409 pKa = 11.18EE410 pKa = 4.53KK411 pKa = 10.7II412 pKa = 3.73
MM1 pKa = 7.61HH2 pKa = 7.1NTTALQSSYY11 pKa = 10.8PNRR14 pKa = 11.84NSRR17 pKa = 11.84SPTANRR23 pKa = 11.84DD24 pKa = 3.09GAIRR28 pKa = 11.84NQSPRR33 pKa = 11.84TQQRR37 pKa = 11.84LSRR40 pKa = 11.84FVFTLNNWTEE50 pKa = 3.8AEE52 pKa = 4.21YY53 pKa = 10.93TDD55 pKa = 5.74IIASDD60 pKa = 4.0VKK62 pKa = 9.59WLVVGKK68 pKa = 10.15EE69 pKa = 3.88IGDD72 pKa = 3.59EE73 pKa = 4.32GTPHH77 pKa = 6.39LQGAVVIGKK86 pKa = 9.26QIAFSTVKK94 pKa = 9.82KK95 pKa = 9.71WPGFSRR101 pKa = 11.84AHH103 pKa = 5.99IEE105 pKa = 3.95NMRR108 pKa = 11.84GKK110 pKa = 9.71PFDD113 pKa = 3.55SLKK116 pKa = 10.9YY117 pKa = 9.23CTKK120 pKa = 10.17QDD122 pKa = 2.72TDD124 pKa = 3.85AFCKK128 pKa = 10.89GEE130 pKa = 4.0MPQEE134 pKa = 4.21GKK136 pKa = 10.82RR137 pKa = 11.84NDD139 pKa = 3.18LHH141 pKa = 7.36DD142 pKa = 5.73AINALKK148 pKa = 10.56QGGTLTSIVKK158 pKa = 10.36SDD160 pKa = 3.96DD161 pKa = 3.27VGLAAAVARR170 pKa = 11.84YY171 pKa = 9.68SKK173 pKa = 10.37GLAMVSSMLKK183 pKa = 9.68PRR185 pKa = 11.84RR186 pKa = 11.84SEE188 pKa = 3.84PPLVIWLSGEE198 pKa = 4.0TGVGKK203 pKa = 8.49TRR205 pKa = 11.84SAVALADD212 pKa = 5.22RR213 pKa = 11.84IAPDD217 pKa = 4.04NYY219 pKa = 9.27WISAGSLRR227 pKa = 11.84WFDD230 pKa = 4.57GYY232 pKa = 10.89DD233 pKa = 3.14GHH235 pKa = 8.5AIAIFDD241 pKa = 4.41DD242 pKa = 4.2LRR244 pKa = 11.84TKK246 pKa = 10.37HH247 pKa = 6.59AEE249 pKa = 3.69FSFLLRR255 pKa = 11.84LLDD258 pKa = 4.3RR259 pKa = 11.84YY260 pKa = 9.37PLRR263 pKa = 11.84VEE265 pKa = 4.48FKK267 pKa = 10.9GGFIDD272 pKa = 3.53WTPSVIVVTAPKK284 pKa = 10.33SPTHH288 pKa = 5.26MWSLRR293 pKa = 11.84NDD295 pKa = 3.27EE296 pKa = 5.6DD297 pKa = 3.61KK298 pKa = 11.24QQLEE302 pKa = 4.26RR303 pKa = 11.84RR304 pKa = 11.84IHH306 pKa = 5.73LAYY309 pKa = 10.36DD310 pKa = 3.44AEE312 pKa = 4.81EE313 pKa = 5.1GDD315 pKa = 3.94DD316 pKa = 4.22WEE318 pKa = 4.78LLLAKK323 pKa = 10.25SEE325 pKa = 4.59AIVAEE330 pKa = 4.54RR331 pKa = 11.84WPTGSYY337 pKa = 8.75PWKK340 pKa = 10.5HH341 pKa = 5.4YY342 pKa = 11.24DD343 pKa = 3.49EE344 pKa = 5.27VNVGSDD350 pKa = 2.93EE351 pKa = 4.17TTIPEE356 pKa = 4.19EE357 pKa = 4.36RR358 pKa = 11.84SCSLSTMGQEE368 pKa = 4.66PDD370 pKa = 3.32TSPSIIEE377 pKa = 4.15IEE379 pKa = 4.05SDD381 pKa = 3.35LSSEE385 pKa = 4.14EE386 pKa = 4.16LVISSSEE393 pKa = 4.07EE394 pKa = 3.53EE395 pKa = 4.18HH396 pKa = 7.74SYY398 pKa = 8.57EE399 pKa = 4.04TQRR402 pKa = 11.84EE403 pKa = 4.34LFSTQYY409 pKa = 11.18EE410 pKa = 4.53KK411 pKa = 10.7II412 pKa = 3.73
Molecular weight: 46.37 kDa
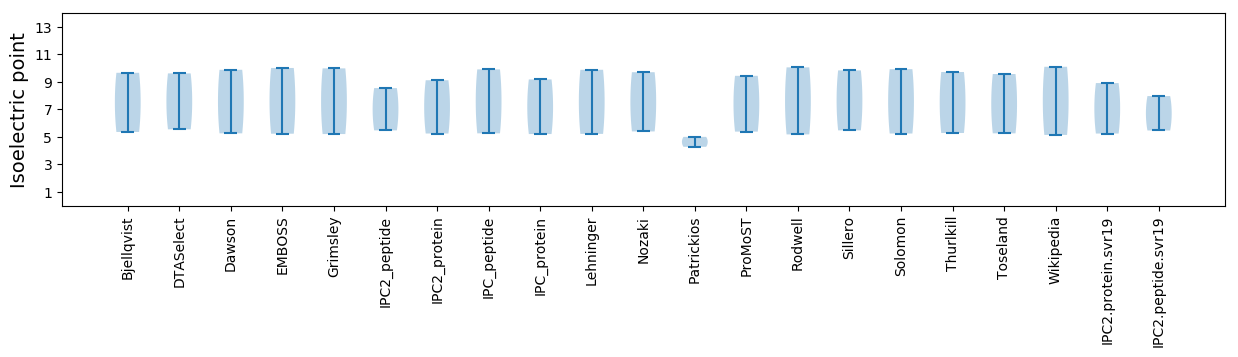
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IB41|A0A0C5IB41_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 15 OX=1618238 PE=3 SV=1
MM1 pKa = 7.71SKK3 pKa = 10.47RR4 pKa = 11.84SYY6 pKa = 11.59NFGTNPSQQYY16 pKa = 9.57GVSRR20 pKa = 11.84ATKK23 pKa = 9.61RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84NVNRR30 pKa = 11.84YY31 pKa = 7.75QNLPRR36 pKa = 11.84GMNQRR41 pKa = 11.84QIIRR45 pKa = 11.84PYY47 pKa = 10.33RR48 pKa = 11.84LYY50 pKa = 10.56TPRR53 pKa = 11.84TPGGQVIAEE62 pKa = 3.82RR63 pKa = 11.84KK64 pKa = 9.91YY65 pKa = 10.91FDD67 pKa = 3.25EE68 pKa = 4.16TLAAKK73 pKa = 9.98IVIAAVTDD81 pKa = 4.06FNDD84 pKa = 3.89AEE86 pKa = 4.11VDD88 pKa = 3.62PAVRR92 pKa = 11.84LCLFAPTQGNDD103 pKa = 2.48ISNRR107 pKa = 11.84EE108 pKa = 3.91SRR110 pKa = 11.84SCFVYY115 pKa = 10.63SITVRR120 pKa = 11.84GSIEE124 pKa = 3.88MLGQGQQTAIDD135 pKa = 3.81NNQIARR141 pKa = 11.84LVLVLDD147 pKa = 3.82KK148 pKa = 10.19QTNGTQMSSEE158 pKa = 4.49DD159 pKa = 4.05LLTTNSGEE167 pKa = 4.22SPMVYY172 pKa = 10.06AYY174 pKa = 11.04QNTANFGRR182 pKa = 11.84FQILKK187 pKa = 9.25DD188 pKa = 3.33QFIEE192 pKa = 4.96FEE194 pKa = 4.12PCNIGGVTGSFWQGGGRR211 pKa = 11.84KK212 pKa = 7.89MFKK215 pKa = 10.22LKK217 pKa = 9.19HH218 pKa = 5.02TFKK221 pKa = 11.06EE222 pKa = 4.37PIRR225 pKa = 11.84VNFGSTNGGTVADD238 pKa = 3.5IVDD241 pKa = 3.62NSFHH245 pKa = 6.97IICGTQNASITTVLNYY261 pKa = 9.91KK262 pKa = 10.18SRR264 pKa = 11.84VSFTGG269 pKa = 3.05
MM1 pKa = 7.71SKK3 pKa = 10.47RR4 pKa = 11.84SYY6 pKa = 11.59NFGTNPSQQYY16 pKa = 9.57GVSRR20 pKa = 11.84ATKK23 pKa = 9.61RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84NVNRR30 pKa = 11.84YY31 pKa = 7.75QNLPRR36 pKa = 11.84GMNQRR41 pKa = 11.84QIIRR45 pKa = 11.84PYY47 pKa = 10.33RR48 pKa = 11.84LYY50 pKa = 10.56TPRR53 pKa = 11.84TPGGQVIAEE62 pKa = 3.82RR63 pKa = 11.84KK64 pKa = 9.91YY65 pKa = 10.91FDD67 pKa = 3.25EE68 pKa = 4.16TLAAKK73 pKa = 9.98IVIAAVTDD81 pKa = 4.06FNDD84 pKa = 3.89AEE86 pKa = 4.11VDD88 pKa = 3.62PAVRR92 pKa = 11.84LCLFAPTQGNDD103 pKa = 2.48ISNRR107 pKa = 11.84EE108 pKa = 3.91SRR110 pKa = 11.84SCFVYY115 pKa = 10.63SITVRR120 pKa = 11.84GSIEE124 pKa = 3.88MLGQGQQTAIDD135 pKa = 3.81NNQIARR141 pKa = 11.84LVLVLDD147 pKa = 3.82KK148 pKa = 10.19QTNGTQMSSEE158 pKa = 4.49DD159 pKa = 4.05LLTTNSGEE167 pKa = 4.22SPMVYY172 pKa = 10.06AYY174 pKa = 11.04QNTANFGRR182 pKa = 11.84FQILKK187 pKa = 9.25DD188 pKa = 3.33QFIEE192 pKa = 4.96FEE194 pKa = 4.12PCNIGGVTGSFWQGGGRR211 pKa = 11.84KK212 pKa = 7.89MFKK215 pKa = 10.22LKK217 pKa = 9.19HH218 pKa = 5.02TFKK221 pKa = 11.06EE222 pKa = 4.37PIRR225 pKa = 11.84VNFGSTNGGTVADD238 pKa = 3.5IVDD241 pKa = 3.62NSFHH245 pKa = 6.97IICGTQNASITTVLNYY261 pKa = 9.91KK262 pKa = 10.18SRR264 pKa = 11.84VSFTGG269 pKa = 3.05
Molecular weight: 30.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
681 |
269 |
412 |
340.5 |
38.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.608 ± 0.56 | 1.028 ± 0.249 |
5.727 ± 0.89 | 6.314 ± 1.411 |
4.112 ± 0.796 | 7.195 ± 0.736 |
1.762 ± 0.553 | 6.167 ± 0.285 |
4.846 ± 0.411 | 6.755 ± 0.842 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.909 ± 0.175 | 4.993 ± 1.529 |
4.405 ± 0.374 | 4.699 ± 1.082 |
6.902 ± 0.492 | 8.664 ± 0.869 |
7.048 ± 0.614 | 5.874 ± 0.444 |
1.762 ± 0.755 | 3.231 ± 0.265 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
