
Rudbeckia flower distortion virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Ruflodivirus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
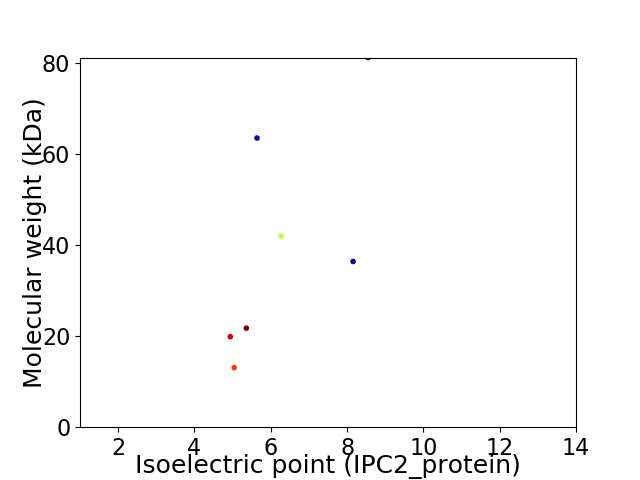
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

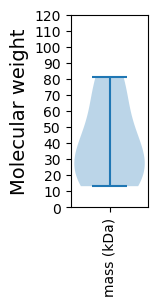
Protein with the lowest isoelectric point:
>tr|B8Y873|B8Y873_9VIRU Uncharacterized protein OS=Rudbeckia flower distortion virus OX=587370 PE=4 SV=1
MM1 pKa = 7.99DD2 pKa = 5.08NDD4 pKa = 3.84FLEE7 pKa = 4.12LLYY10 pKa = 10.83RR11 pKa = 11.84NKK13 pKa = 10.16QPLCEE18 pKa = 4.15NNAIRR23 pKa = 11.84YY24 pKa = 7.75ANKK27 pKa = 9.73VDD29 pKa = 3.81DD30 pKa = 4.25LLYY33 pKa = 9.39YY34 pKa = 10.28QEE36 pKa = 4.54FLVLKK41 pKa = 10.53AIDD44 pKa = 3.74EE45 pKa = 4.96LIFMRR50 pKa = 11.84NCQQTYY56 pKa = 7.69NQVYY60 pKa = 10.67SRR62 pKa = 11.84GSDD65 pKa = 3.19LNRR68 pKa = 11.84FYY70 pKa = 11.5GLQLEE75 pKa = 4.46SEE77 pKa = 4.39IKK79 pKa = 9.95IEE81 pKa = 3.92KK82 pKa = 10.62LEE84 pKa = 4.81DD85 pKa = 3.02IRR87 pKa = 11.84KK88 pKa = 8.1QFLMHH93 pKa = 7.06SDD95 pKa = 3.36NNYY98 pKa = 9.9CLHH101 pKa = 6.49NNLPRR106 pKa = 11.84GVQPPITSVYY116 pKa = 10.94NGTTTFKK123 pKa = 11.22GNYY126 pKa = 8.22SEE128 pKa = 6.12NYY130 pKa = 9.61LCSLDD135 pKa = 3.58SRR137 pKa = 11.84IVEE140 pKa = 4.16KK141 pKa = 10.58RR142 pKa = 11.84SKK144 pKa = 10.57EE145 pKa = 3.93LLEE148 pKa = 4.12LRR150 pKa = 11.84QALSEE155 pKa = 4.21SLPLLSSSEE164 pKa = 4.53DD165 pKa = 3.67EE166 pKa = 4.69NSDD169 pKa = 3.25
MM1 pKa = 7.99DD2 pKa = 5.08NDD4 pKa = 3.84FLEE7 pKa = 4.12LLYY10 pKa = 10.83RR11 pKa = 11.84NKK13 pKa = 10.16QPLCEE18 pKa = 4.15NNAIRR23 pKa = 11.84YY24 pKa = 7.75ANKK27 pKa = 9.73VDD29 pKa = 3.81DD30 pKa = 4.25LLYY33 pKa = 9.39YY34 pKa = 10.28QEE36 pKa = 4.54FLVLKK41 pKa = 10.53AIDD44 pKa = 3.74EE45 pKa = 4.96LIFMRR50 pKa = 11.84NCQQTYY56 pKa = 7.69NQVYY60 pKa = 10.67SRR62 pKa = 11.84GSDD65 pKa = 3.19LNRR68 pKa = 11.84FYY70 pKa = 11.5GLQLEE75 pKa = 4.46SEE77 pKa = 4.39IKK79 pKa = 9.95IEE81 pKa = 3.92KK82 pKa = 10.62LEE84 pKa = 4.81DD85 pKa = 3.02IRR87 pKa = 11.84KK88 pKa = 8.1QFLMHH93 pKa = 7.06SDD95 pKa = 3.36NNYY98 pKa = 9.9CLHH101 pKa = 6.49NNLPRR106 pKa = 11.84GVQPPITSVYY116 pKa = 10.94NGTTTFKK123 pKa = 11.22GNYY126 pKa = 8.22SEE128 pKa = 6.12NYY130 pKa = 9.61LCSLDD135 pKa = 3.58SRR137 pKa = 11.84IVEE140 pKa = 4.16KK141 pKa = 10.58RR142 pKa = 11.84SKK144 pKa = 10.57EE145 pKa = 3.93LLEE148 pKa = 4.12LRR150 pKa = 11.84QALSEE155 pKa = 4.21SLPLLSSSEE164 pKa = 4.53DD165 pKa = 3.67EE166 pKa = 4.69NSDD169 pKa = 3.25
Molecular weight: 19.87 kDa
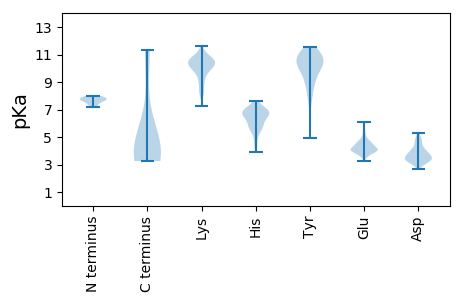
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8Y872|B8Y872_9VIRU Putative translational transactivator OS=Rudbeckia flower distortion virus OX=587370 PE=4 SV=1
MM1 pKa = 7.2TKK3 pKa = 10.51SKK5 pKa = 9.04ITLLMNLNLKK15 pKa = 9.37TSGKK19 pKa = 7.24STKK22 pKa = 8.93TNPYY26 pKa = 7.48STYY29 pKa = 8.96VTVGFYY35 pKa = 10.8FNGYY39 pKa = 9.92KK40 pKa = 10.38GFHH43 pKa = 5.58LHH45 pKa = 7.14AYY47 pKa = 9.05VDD49 pKa = 4.05TGATSCTASPHH60 pKa = 6.6IIPTEE65 pKa = 3.69LWEE68 pKa = 4.35EE69 pKa = 4.09LPKK72 pKa = 10.63PILVNIANDD81 pKa = 3.6TNIEE85 pKa = 3.62IRR87 pKa = 11.84YY88 pKa = 4.93VTRR91 pKa = 11.84NLKK94 pKa = 10.53VSMTDD99 pKa = 3.14TNGNKK104 pKa = 8.34HH105 pKa = 5.47TFIIPTVYY113 pKa = 10.07QQNTGMDD120 pKa = 4.44FIFGQNFLKK129 pKa = 10.66LYY131 pKa = 10.68RR132 pKa = 11.84PFTQDD137 pKa = 2.75LYY139 pKa = 11.2EE140 pKa = 5.49IYY142 pKa = 9.54LTPKK146 pKa = 9.93LLKK149 pKa = 10.55FKK151 pKa = 10.14IQQRR155 pKa = 11.84AFKK158 pKa = 9.83QASPGFLDD166 pKa = 4.86SIRR169 pKa = 11.84KK170 pKa = 7.55QKK172 pKa = 10.45RR173 pKa = 11.84GEE175 pKa = 4.33IISKK179 pKa = 10.05HH180 pKa = 4.48ATNPINITKK189 pKa = 10.56LEE191 pKa = 4.55DD192 pKa = 3.14NLRR195 pKa = 11.84IVEE198 pKa = 4.63TILEE202 pKa = 4.34DD203 pKa = 4.05SNSKK207 pKa = 8.12TLSEE211 pKa = 4.26VSDD214 pKa = 3.81SKK216 pKa = 10.19TLSEE220 pKa = 5.32DD221 pKa = 3.07SDD223 pKa = 4.68DD224 pKa = 3.97YY225 pKa = 11.25FVYY228 pKa = 10.61SIQRR232 pKa = 11.84HH233 pKa = 3.93NTIVNDD239 pKa = 4.82LLTQACSEE247 pKa = 4.24NPLDD251 pKa = 4.61EE252 pKa = 4.8NKK254 pKa = 10.14NHH256 pKa = 6.6NGLLAEE262 pKa = 4.72IKK264 pKa = 10.24LINPATTVNVKK275 pKa = 10.52SMAYY279 pKa = 10.06SPDD282 pKa = 3.41DD283 pKa = 3.31AVEE286 pKa = 3.99INKK289 pKa = 9.6QIQEE293 pKa = 4.1LLEE296 pKa = 4.18IKK298 pKa = 10.21VIRR301 pKa = 11.84PSRR304 pKa = 11.84SPHH307 pKa = 6.12SSPCFLVQNHH317 pKa = 5.5NEE319 pKa = 3.64IKK321 pKa = 10.12RR322 pKa = 11.84GKK324 pKa = 9.2KK325 pKa = 9.74RR326 pKa = 11.84LVINYY331 pKa = 7.48KK332 pKa = 10.2ALNAATISDD341 pKa = 4.65GYY343 pKa = 11.09LLPNKK348 pKa = 8.19EE349 pKa = 4.66TILTAIRR356 pKa = 11.84GRR358 pKa = 11.84KK359 pKa = 8.78YY360 pKa = 10.8FSTLDD365 pKa = 3.71CKK367 pKa = 11.09SGFWQIRR374 pKa = 11.84LNEE377 pKa = 3.95NSKK380 pKa = 10.32PLTAFSCPMGQYY392 pKa = 8.48EE393 pKa = 4.45WNVVPFGLKK402 pKa = 9.09QAPGLFQRR410 pKa = 11.84FMDD413 pKa = 3.83NSFKK417 pKa = 10.7EE418 pKa = 4.26YY419 pKa = 10.71SAFCAVYY426 pKa = 10.66VDD428 pKa = 5.31DD429 pKa = 5.26ILVFSKK435 pKa = 9.67TLDD438 pKa = 3.12EE439 pKa = 5.43HH440 pKa = 6.83YY441 pKa = 11.32DD442 pKa = 3.4HH443 pKa = 7.38LEE445 pKa = 3.87TVLRR449 pKa = 11.84KK450 pKa = 10.08CIEE453 pKa = 3.87TGIILSKK460 pKa = 10.71KK461 pKa = 8.14KK462 pKa = 10.95AEE464 pKa = 4.09VAKK467 pKa = 10.16TKK469 pKa = 10.4INYY472 pKa = 10.01LGFTISNGEE481 pKa = 3.95IEE483 pKa = 4.72LQSHH487 pKa = 5.67ILEE490 pKa = 4.69NIKK493 pKa = 10.42LFPSRR498 pKa = 11.84IPDD501 pKa = 3.34KK502 pKa = 11.02KK503 pKa = 10.4SLQRR507 pKa = 11.84FLGILTYY514 pKa = 10.83ADD516 pKa = 3.09QYY518 pKa = 10.68IRR520 pKa = 11.84KK521 pKa = 8.48LAEE524 pKa = 3.4WRR526 pKa = 11.84KK527 pKa = 8.54PLQRR531 pKa = 11.84KK532 pKa = 8.35LKK534 pKa = 10.37KK535 pKa = 9.77DD536 pKa = 4.01TVWEE540 pKa = 4.32WNDD543 pKa = 3.07SDD545 pKa = 3.7TQYY548 pKa = 11.32VEE550 pKa = 3.78KK551 pKa = 10.62FKK553 pKa = 11.33RR554 pKa = 11.84NLKK557 pKa = 9.52EE558 pKa = 3.81FPKK561 pKa = 10.27LHH563 pKa = 6.64HH564 pKa = 6.88PLPDD568 pKa = 3.22EE569 pKa = 4.02YY570 pKa = 11.37LIIEE574 pKa = 4.42TDD576 pKa = 3.07ASHH579 pKa = 5.86EE580 pKa = 4.04HH581 pKa = 5.8WAGVLKK587 pKa = 11.01SRR589 pKa = 11.84GTDD592 pKa = 3.06NLEE595 pKa = 3.65RR596 pKa = 11.84LCRR599 pKa = 11.84YY600 pKa = 7.89TSGSFKK606 pKa = 10.2PAEE609 pKa = 4.13INYY612 pKa = 9.4HH613 pKa = 4.93SNEE616 pKa = 4.07KK617 pKa = 10.24EE618 pKa = 3.87VLAVKK623 pKa = 9.46RR624 pKa = 11.84TITKK628 pKa = 9.96FKK630 pKa = 10.6GYY632 pKa = 9.46LASSEE637 pKa = 4.08FLVRR641 pKa = 11.84TDD643 pKa = 3.37NKK645 pKa = 10.13YY646 pKa = 8.35FTYY649 pKa = 10.6FLRR652 pKa = 11.84TSIKK656 pKa = 10.45DD657 pKa = 3.29DD658 pKa = 3.67YY659 pKa = 11.06KK660 pKa = 10.54QGRR663 pKa = 11.84LIRR666 pKa = 11.84WQQWFSHH673 pKa = 4.93YY674 pKa = 10.35KK675 pKa = 10.32FNVEE679 pKa = 4.28HH680 pKa = 7.03LAGTHH685 pKa = 6.09NFTADD690 pKa = 2.87SLTRR694 pKa = 11.84EE695 pKa = 4.62FANKK699 pKa = 9.67PPP701 pKa = 3.82
MM1 pKa = 7.2TKK3 pKa = 10.51SKK5 pKa = 9.04ITLLMNLNLKK15 pKa = 9.37TSGKK19 pKa = 7.24STKK22 pKa = 8.93TNPYY26 pKa = 7.48STYY29 pKa = 8.96VTVGFYY35 pKa = 10.8FNGYY39 pKa = 9.92KK40 pKa = 10.38GFHH43 pKa = 5.58LHH45 pKa = 7.14AYY47 pKa = 9.05VDD49 pKa = 4.05TGATSCTASPHH60 pKa = 6.6IIPTEE65 pKa = 3.69LWEE68 pKa = 4.35EE69 pKa = 4.09LPKK72 pKa = 10.63PILVNIANDD81 pKa = 3.6TNIEE85 pKa = 3.62IRR87 pKa = 11.84YY88 pKa = 4.93VTRR91 pKa = 11.84NLKK94 pKa = 10.53VSMTDD99 pKa = 3.14TNGNKK104 pKa = 8.34HH105 pKa = 5.47TFIIPTVYY113 pKa = 10.07QQNTGMDD120 pKa = 4.44FIFGQNFLKK129 pKa = 10.66LYY131 pKa = 10.68RR132 pKa = 11.84PFTQDD137 pKa = 2.75LYY139 pKa = 11.2EE140 pKa = 5.49IYY142 pKa = 9.54LTPKK146 pKa = 9.93LLKK149 pKa = 10.55FKK151 pKa = 10.14IQQRR155 pKa = 11.84AFKK158 pKa = 9.83QASPGFLDD166 pKa = 4.86SIRR169 pKa = 11.84KK170 pKa = 7.55QKK172 pKa = 10.45RR173 pKa = 11.84GEE175 pKa = 4.33IISKK179 pKa = 10.05HH180 pKa = 4.48ATNPINITKK189 pKa = 10.56LEE191 pKa = 4.55DD192 pKa = 3.14NLRR195 pKa = 11.84IVEE198 pKa = 4.63TILEE202 pKa = 4.34DD203 pKa = 4.05SNSKK207 pKa = 8.12TLSEE211 pKa = 4.26VSDD214 pKa = 3.81SKK216 pKa = 10.19TLSEE220 pKa = 5.32DD221 pKa = 3.07SDD223 pKa = 4.68DD224 pKa = 3.97YY225 pKa = 11.25FVYY228 pKa = 10.61SIQRR232 pKa = 11.84HH233 pKa = 3.93NTIVNDD239 pKa = 4.82LLTQACSEE247 pKa = 4.24NPLDD251 pKa = 4.61EE252 pKa = 4.8NKK254 pKa = 10.14NHH256 pKa = 6.6NGLLAEE262 pKa = 4.72IKK264 pKa = 10.24LINPATTVNVKK275 pKa = 10.52SMAYY279 pKa = 10.06SPDD282 pKa = 3.41DD283 pKa = 3.31AVEE286 pKa = 3.99INKK289 pKa = 9.6QIQEE293 pKa = 4.1LLEE296 pKa = 4.18IKK298 pKa = 10.21VIRR301 pKa = 11.84PSRR304 pKa = 11.84SPHH307 pKa = 6.12SSPCFLVQNHH317 pKa = 5.5NEE319 pKa = 3.64IKK321 pKa = 10.12RR322 pKa = 11.84GKK324 pKa = 9.2KK325 pKa = 9.74RR326 pKa = 11.84LVINYY331 pKa = 7.48KK332 pKa = 10.2ALNAATISDD341 pKa = 4.65GYY343 pKa = 11.09LLPNKK348 pKa = 8.19EE349 pKa = 4.66TILTAIRR356 pKa = 11.84GRR358 pKa = 11.84KK359 pKa = 8.78YY360 pKa = 10.8FSTLDD365 pKa = 3.71CKK367 pKa = 11.09SGFWQIRR374 pKa = 11.84LNEE377 pKa = 3.95NSKK380 pKa = 10.32PLTAFSCPMGQYY392 pKa = 8.48EE393 pKa = 4.45WNVVPFGLKK402 pKa = 9.09QAPGLFQRR410 pKa = 11.84FMDD413 pKa = 3.83NSFKK417 pKa = 10.7EE418 pKa = 4.26YY419 pKa = 10.71SAFCAVYY426 pKa = 10.66VDD428 pKa = 5.31DD429 pKa = 5.26ILVFSKK435 pKa = 9.67TLDD438 pKa = 3.12EE439 pKa = 5.43HH440 pKa = 6.83YY441 pKa = 11.32DD442 pKa = 3.4HH443 pKa = 7.38LEE445 pKa = 3.87TVLRR449 pKa = 11.84KK450 pKa = 10.08CIEE453 pKa = 3.87TGIILSKK460 pKa = 10.71KK461 pKa = 8.14KK462 pKa = 10.95AEE464 pKa = 4.09VAKK467 pKa = 10.16TKK469 pKa = 10.4INYY472 pKa = 10.01LGFTISNGEE481 pKa = 3.95IEE483 pKa = 4.72LQSHH487 pKa = 5.67ILEE490 pKa = 4.69NIKK493 pKa = 10.42LFPSRR498 pKa = 11.84IPDD501 pKa = 3.34KK502 pKa = 11.02KK503 pKa = 10.4SLQRR507 pKa = 11.84FLGILTYY514 pKa = 10.83ADD516 pKa = 3.09QYY518 pKa = 10.68IRR520 pKa = 11.84KK521 pKa = 8.48LAEE524 pKa = 3.4WRR526 pKa = 11.84KK527 pKa = 8.54PLQRR531 pKa = 11.84KK532 pKa = 8.35LKK534 pKa = 10.37KK535 pKa = 9.77DD536 pKa = 4.01TVWEE540 pKa = 4.32WNDD543 pKa = 3.07SDD545 pKa = 3.7TQYY548 pKa = 11.32VEE550 pKa = 3.78KK551 pKa = 10.62FKK553 pKa = 11.33RR554 pKa = 11.84NLKK557 pKa = 9.52EE558 pKa = 3.81FPKK561 pKa = 10.27LHH563 pKa = 6.64HH564 pKa = 6.88PLPDD568 pKa = 3.22EE569 pKa = 4.02YY570 pKa = 11.37LIIEE574 pKa = 4.42TDD576 pKa = 3.07ASHH579 pKa = 5.86EE580 pKa = 4.04HH581 pKa = 5.8WAGVLKK587 pKa = 11.01SRR589 pKa = 11.84GTDD592 pKa = 3.06NLEE595 pKa = 3.65RR596 pKa = 11.84LCRR599 pKa = 11.84YY600 pKa = 7.89TSGSFKK606 pKa = 10.2PAEE609 pKa = 4.13INYY612 pKa = 9.4HH613 pKa = 4.93SNEE616 pKa = 4.07KK617 pKa = 10.24EE618 pKa = 3.87VLAVKK623 pKa = 9.46RR624 pKa = 11.84TITKK628 pKa = 9.96FKK630 pKa = 10.6GYY632 pKa = 9.46LASSEE637 pKa = 4.08FLVRR641 pKa = 11.84TDD643 pKa = 3.37NKK645 pKa = 10.13YY646 pKa = 8.35FTYY649 pKa = 10.6FLRR652 pKa = 11.84TSIKK656 pKa = 10.45DD657 pKa = 3.29DD658 pKa = 3.67YY659 pKa = 11.06KK660 pKa = 10.54QGRR663 pKa = 11.84LIRR666 pKa = 11.84WQQWFSHH673 pKa = 4.93YY674 pKa = 10.35KK675 pKa = 10.32FNVEE679 pKa = 4.28HH680 pKa = 7.03LAGTHH685 pKa = 6.09NFTADD690 pKa = 2.87SLTRR694 pKa = 11.84EE695 pKa = 4.62FANKK699 pKa = 9.67PPP701 pKa = 3.82
Molecular weight: 81.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2397 |
112 |
701 |
342.4 |
39.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.297 ± 0.631 | 1.585 ± 0.333 |
6.008 ± 0.352 | 7.468 ± 0.774 |
4.422 ± 0.344 | 3.504 ± 0.292 |
2.336 ± 0.264 | 7.968 ± 0.888 |
8.761 ± 0.498 | 9.595 ± 0.79 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.418 ± 0.311 | 7.134 ± 0.408 |
4.172 ± 0.345 | 4.172 ± 0.558 |
4.631 ± 0.497 | 7.217 ± 0.416 |
6.258 ± 0.606 | 3.963 ± 0.21 |
0.876 ± 0.185 | 4.214 ± 0.376 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
