
Caprine arthritis encephalitis virus Ov496
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Lentivirus; Caprine arthritis encephalitis virus
Average proteome isoelectric point is 8.59
Get precalculated fractions of proteins
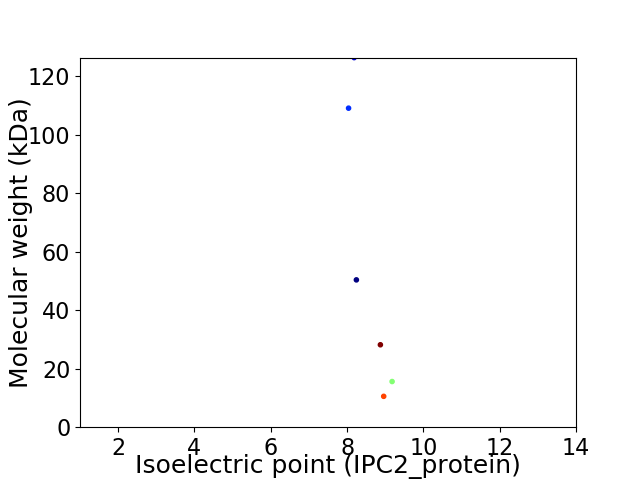
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0JIH3|C0JIH3_CAEV Rev protein OS=Caprine arthritis encephalitis virus Ov496 OX=621214 PE=4 SV=1
MM1 pKa = 8.17DD2 pKa = 4.16MGAKK6 pKa = 9.9HH7 pKa = 5.43MQRR10 pKa = 11.84TGEE13 pKa = 4.25GNWVEE18 pKa = 4.28VKK20 pKa = 9.96MEE22 pKa = 3.88EE23 pKa = 4.24QEE25 pKa = 4.47RR26 pKa = 11.84EE27 pKa = 4.21EE28 pKa = 4.35EE29 pKa = 4.21PLLHH33 pKa = 6.36TRR35 pKa = 11.84QQGKK39 pKa = 9.67YY40 pKa = 8.4KK41 pKa = 9.98PQVSEE46 pKa = 4.12QILNGDD52 pKa = 3.76TNTCLTYY59 pKa = 10.96AWIFLWGIQLTLWILLLLQWPSMGCKK85 pKa = 10.18AEE87 pKa = 4.88DD88 pKa = 4.3YY89 pKa = 8.67ITLISDD95 pKa = 4.17PYY97 pKa = 10.58GFQPVRR103 pKa = 11.84NVSGVPVTCVTKK115 pKa = 10.79KK116 pKa = 9.06FAKK119 pKa = 9.41WGCQPIGAYY128 pKa = 8.56PNPEE132 pKa = 3.21IEE134 pKa = 4.19YY135 pKa = 10.4RR136 pKa = 11.84NISSDD141 pKa = 2.93ILKK144 pKa = 10.21EE145 pKa = 4.27VYY147 pKa = 10.14QEE149 pKa = 3.69DD150 pKa = 4.59WPWNTYY156 pKa = 8.43HH157 pKa = 6.52WPLWQMEE164 pKa = 4.21NVKK167 pKa = 9.81QWLKK171 pKa = 11.05EE172 pKa = 3.78NEE174 pKa = 3.74KK175 pKa = 10.78DD176 pKa = 3.98YY177 pKa = 11.22KK178 pKa = 10.8RR179 pKa = 11.84KK180 pKa = 10.45LNTTKK185 pKa = 10.93EE186 pKa = 4.15DD187 pKa = 3.84LDD189 pKa = 3.73EE190 pKa = 4.58LLAGKK195 pKa = 9.81IRR197 pKa = 11.84GRR199 pKa = 11.84FCVPYY204 pKa = 10.13PFALLKK210 pKa = 9.28CTEE213 pKa = 4.18WCWYY217 pKa = 8.3PAAIDD222 pKa = 4.31DD223 pKa = 4.03EE224 pKa = 4.76SGRR227 pKa = 11.84AKK229 pKa = 10.44NIKK232 pKa = 9.63INCTNARR239 pKa = 11.84AVSCTEE245 pKa = 3.7EE246 pKa = 4.04MPLASIHH253 pKa = 5.62RR254 pKa = 11.84AYY256 pKa = 8.95WGKK259 pKa = 10.3RR260 pKa = 11.84EE261 pKa = 3.98RR262 pKa = 11.84EE263 pKa = 3.99SMQFMNIRR271 pKa = 11.84ACQGSKK277 pKa = 10.19LRR279 pKa = 11.84CQRR282 pKa = 11.84RR283 pKa = 11.84PGGCMEE289 pKa = 5.78GYY291 pKa = 9.02PIPIGAEE298 pKa = 3.92IIPEE302 pKa = 3.97SMKK305 pKa = 10.57YY306 pKa = 10.55LRR308 pKa = 11.84GKK310 pKa = 9.48KK311 pKa = 8.91SQYY314 pKa = 11.07GGIKK318 pKa = 9.98DD319 pKa = 3.82RR320 pKa = 11.84NGEE323 pKa = 4.28LNLPLTVRR331 pKa = 11.84VWVKK335 pKa = 10.19LANNSAWINGAPPHH349 pKa = 5.93WKK351 pKa = 10.21DD352 pKa = 3.78RR353 pKa = 11.84IKK355 pKa = 10.81GSKK358 pKa = 9.59GINGTLWGEE367 pKa = 4.29LKK369 pKa = 10.6SIHH372 pKa = 6.56HH373 pKa = 6.85LGFAVSYY380 pKa = 10.31DD381 pKa = 3.7GKK383 pKa = 8.85WCNFTKK389 pKa = 10.38HH390 pKa = 5.64IRR392 pKa = 11.84IGQEE396 pKa = 3.45VFPYY400 pKa = 9.92HH401 pKa = 6.63YY402 pKa = 9.95KK403 pKa = 10.68PSWNCSAAKK412 pKa = 6.77WTQYY416 pKa = 7.78PTWQVTRR423 pKa = 11.84YY424 pKa = 9.86LDD426 pKa = 3.71MVEE429 pKa = 5.02HH430 pKa = 5.5MTGEE434 pKa = 4.43CVQRR438 pKa = 11.84PEE440 pKa = 3.89RR441 pKa = 11.84HH442 pKa = 6.27NITVGNGTITGNCSVTNWDD461 pKa = 3.46GCNCSRR467 pKa = 11.84SGNHH471 pKa = 6.9LYY473 pKa = 10.69NSSSGGLLVIICRR486 pKa = 11.84QNKK489 pKa = 8.54TITGIMGTNINWTTMMGILKK509 pKa = 9.94NCSGCGNATLDD520 pKa = 3.64RR521 pKa = 11.84TGTGALGNVKK531 pKa = 10.28NEE533 pKa = 4.08NCSLPHH539 pKa = 7.55INEE542 pKa = 4.29SNKK545 pKa = 7.61WTCAPRR551 pKa = 11.84WRR553 pKa = 11.84GGMSDD558 pKa = 3.24SLYY561 pKa = 10.52IAGGKK566 pKa = 7.62QFWTRR571 pKa = 11.84VKK573 pKa = 10.49AQYY576 pKa = 10.37SCEE579 pKa = 4.29SNLEE583 pKa = 4.05EE584 pKa = 5.32LDD586 pKa = 4.18GMMHH590 pKa = 5.97QQILLQKK597 pKa = 8.19YY598 pKa = 6.7QVIRR602 pKa = 11.84VRR604 pKa = 11.84AYY606 pKa = 7.72TYY608 pKa = 11.42GMIDD612 pKa = 3.54MPSSHH617 pKa = 6.04NQRR620 pKa = 11.84STRR623 pKa = 11.84RR624 pKa = 11.84KK625 pKa = 9.06RR626 pKa = 11.84EE627 pKa = 3.6LGHH630 pKa = 6.04RR631 pKa = 11.84RR632 pKa = 11.84KK633 pKa = 10.09KK634 pKa = 10.55RR635 pKa = 11.84GIGLVIMLVTMAIVAAAGASLGVANAIQQSYY666 pKa = 8.18TKK668 pKa = 10.67AAVQTLANATAAQQDD683 pKa = 4.19VLEE686 pKa = 4.4ATYY689 pKa = 11.78AMVQHH694 pKa = 5.93VAKK697 pKa = 10.35GVRR700 pKa = 11.84ILEE703 pKa = 4.04ARR705 pKa = 11.84VARR708 pKa = 11.84VEE710 pKa = 4.57AITDD714 pKa = 4.42RR715 pKa = 11.84IMLYY719 pKa = 10.18QEE721 pKa = 4.25LDD723 pKa = 3.1CWHH726 pKa = 5.77YY727 pKa = 10.7HH728 pKa = 4.7QYY730 pKa = 11.15CVTSTRR736 pKa = 11.84AEE738 pKa = 3.88VAKK741 pKa = 10.3YY742 pKa = 10.58INWTRR747 pKa = 11.84FKK749 pKa = 11.2DD750 pKa = 3.51NCTWQQWEE758 pKa = 4.66RR759 pKa = 11.84EE760 pKa = 4.31LQGHH764 pKa = 6.37DD765 pKa = 3.84EE766 pKa = 4.32NLTMLLRR773 pKa = 11.84EE774 pKa = 4.5SARR777 pKa = 11.84MTQLAQEE784 pKa = 4.21QARR787 pKa = 11.84RR788 pKa = 11.84IPDD791 pKa = 2.85VWEE794 pKa = 4.19ALQQVFDD801 pKa = 3.76WSGWFSWLKK810 pKa = 9.6YY811 pKa = 9.94IPLIVIGILGCIIIRR826 pKa = 11.84IMICVCQPLIQVYY839 pKa = 7.94RR840 pKa = 11.84TLSTPRR846 pKa = 11.84YY847 pKa = 8.51QKK849 pKa = 9.22VTVIMEE855 pKa = 4.04TRR857 pKa = 11.84ADD859 pKa = 3.56VGGEE863 pKa = 3.74NAQDD867 pKa = 3.42FGNGFEE873 pKa = 4.1EE874 pKa = 5.02LEE876 pKa = 4.36ISRR879 pKa = 11.84EE880 pKa = 4.03PQKK883 pKa = 10.38VTIKK887 pKa = 10.54KK888 pKa = 8.57AWSNAWEE895 pKa = 4.11LWEE898 pKa = 3.92TSPWEE903 pKa = 4.05EE904 pKa = 3.25RR905 pKa = 11.84WKK907 pKa = 11.21KK908 pKa = 10.56NLLKK912 pKa = 10.42PLILPLTMGIWINGLLGEE930 pKa = 4.06RR931 pKa = 11.84HH932 pKa = 5.71KK933 pKa = 11.24NKK935 pKa = 10.06KK936 pKa = 9.39EE937 pKa = 3.97RR938 pKa = 11.84VDD940 pKa = 3.57CEE942 pKa = 3.84TWEE945 pKa = 4.58KK946 pKa = 11.38GDD948 pKa = 3.53
MM1 pKa = 8.17DD2 pKa = 4.16MGAKK6 pKa = 9.9HH7 pKa = 5.43MQRR10 pKa = 11.84TGEE13 pKa = 4.25GNWVEE18 pKa = 4.28VKK20 pKa = 9.96MEE22 pKa = 3.88EE23 pKa = 4.24QEE25 pKa = 4.47RR26 pKa = 11.84EE27 pKa = 4.21EE28 pKa = 4.35EE29 pKa = 4.21PLLHH33 pKa = 6.36TRR35 pKa = 11.84QQGKK39 pKa = 9.67YY40 pKa = 8.4KK41 pKa = 9.98PQVSEE46 pKa = 4.12QILNGDD52 pKa = 3.76TNTCLTYY59 pKa = 10.96AWIFLWGIQLTLWILLLLQWPSMGCKK85 pKa = 10.18AEE87 pKa = 4.88DD88 pKa = 4.3YY89 pKa = 8.67ITLISDD95 pKa = 4.17PYY97 pKa = 10.58GFQPVRR103 pKa = 11.84NVSGVPVTCVTKK115 pKa = 10.79KK116 pKa = 9.06FAKK119 pKa = 9.41WGCQPIGAYY128 pKa = 8.56PNPEE132 pKa = 3.21IEE134 pKa = 4.19YY135 pKa = 10.4RR136 pKa = 11.84NISSDD141 pKa = 2.93ILKK144 pKa = 10.21EE145 pKa = 4.27VYY147 pKa = 10.14QEE149 pKa = 3.69DD150 pKa = 4.59WPWNTYY156 pKa = 8.43HH157 pKa = 6.52WPLWQMEE164 pKa = 4.21NVKK167 pKa = 9.81QWLKK171 pKa = 11.05EE172 pKa = 3.78NEE174 pKa = 3.74KK175 pKa = 10.78DD176 pKa = 3.98YY177 pKa = 11.22KK178 pKa = 10.8RR179 pKa = 11.84KK180 pKa = 10.45LNTTKK185 pKa = 10.93EE186 pKa = 4.15DD187 pKa = 3.84LDD189 pKa = 3.73EE190 pKa = 4.58LLAGKK195 pKa = 9.81IRR197 pKa = 11.84GRR199 pKa = 11.84FCVPYY204 pKa = 10.13PFALLKK210 pKa = 9.28CTEE213 pKa = 4.18WCWYY217 pKa = 8.3PAAIDD222 pKa = 4.31DD223 pKa = 4.03EE224 pKa = 4.76SGRR227 pKa = 11.84AKK229 pKa = 10.44NIKK232 pKa = 9.63INCTNARR239 pKa = 11.84AVSCTEE245 pKa = 3.7EE246 pKa = 4.04MPLASIHH253 pKa = 5.62RR254 pKa = 11.84AYY256 pKa = 8.95WGKK259 pKa = 10.3RR260 pKa = 11.84EE261 pKa = 3.98RR262 pKa = 11.84EE263 pKa = 3.99SMQFMNIRR271 pKa = 11.84ACQGSKK277 pKa = 10.19LRR279 pKa = 11.84CQRR282 pKa = 11.84RR283 pKa = 11.84PGGCMEE289 pKa = 5.78GYY291 pKa = 9.02PIPIGAEE298 pKa = 3.92IIPEE302 pKa = 3.97SMKK305 pKa = 10.57YY306 pKa = 10.55LRR308 pKa = 11.84GKK310 pKa = 9.48KK311 pKa = 8.91SQYY314 pKa = 11.07GGIKK318 pKa = 9.98DD319 pKa = 3.82RR320 pKa = 11.84NGEE323 pKa = 4.28LNLPLTVRR331 pKa = 11.84VWVKK335 pKa = 10.19LANNSAWINGAPPHH349 pKa = 5.93WKK351 pKa = 10.21DD352 pKa = 3.78RR353 pKa = 11.84IKK355 pKa = 10.81GSKK358 pKa = 9.59GINGTLWGEE367 pKa = 4.29LKK369 pKa = 10.6SIHH372 pKa = 6.56HH373 pKa = 6.85LGFAVSYY380 pKa = 10.31DD381 pKa = 3.7GKK383 pKa = 8.85WCNFTKK389 pKa = 10.38HH390 pKa = 5.64IRR392 pKa = 11.84IGQEE396 pKa = 3.45VFPYY400 pKa = 9.92HH401 pKa = 6.63YY402 pKa = 9.95KK403 pKa = 10.68PSWNCSAAKK412 pKa = 6.77WTQYY416 pKa = 7.78PTWQVTRR423 pKa = 11.84YY424 pKa = 9.86LDD426 pKa = 3.71MVEE429 pKa = 5.02HH430 pKa = 5.5MTGEE434 pKa = 4.43CVQRR438 pKa = 11.84PEE440 pKa = 3.89RR441 pKa = 11.84HH442 pKa = 6.27NITVGNGTITGNCSVTNWDD461 pKa = 3.46GCNCSRR467 pKa = 11.84SGNHH471 pKa = 6.9LYY473 pKa = 10.69NSSSGGLLVIICRR486 pKa = 11.84QNKK489 pKa = 8.54TITGIMGTNINWTTMMGILKK509 pKa = 9.94NCSGCGNATLDD520 pKa = 3.64RR521 pKa = 11.84TGTGALGNVKK531 pKa = 10.28NEE533 pKa = 4.08NCSLPHH539 pKa = 7.55INEE542 pKa = 4.29SNKK545 pKa = 7.61WTCAPRR551 pKa = 11.84WRR553 pKa = 11.84GGMSDD558 pKa = 3.24SLYY561 pKa = 10.52IAGGKK566 pKa = 7.62QFWTRR571 pKa = 11.84VKK573 pKa = 10.49AQYY576 pKa = 10.37SCEE579 pKa = 4.29SNLEE583 pKa = 4.05EE584 pKa = 5.32LDD586 pKa = 4.18GMMHH590 pKa = 5.97QQILLQKK597 pKa = 8.19YY598 pKa = 6.7QVIRR602 pKa = 11.84VRR604 pKa = 11.84AYY606 pKa = 7.72TYY608 pKa = 11.42GMIDD612 pKa = 3.54MPSSHH617 pKa = 6.04NQRR620 pKa = 11.84STRR623 pKa = 11.84RR624 pKa = 11.84KK625 pKa = 9.06RR626 pKa = 11.84EE627 pKa = 3.6LGHH630 pKa = 6.04RR631 pKa = 11.84RR632 pKa = 11.84KK633 pKa = 10.09KK634 pKa = 10.55RR635 pKa = 11.84GIGLVIMLVTMAIVAAAGASLGVANAIQQSYY666 pKa = 8.18TKK668 pKa = 10.67AAVQTLANATAAQQDD683 pKa = 4.19VLEE686 pKa = 4.4ATYY689 pKa = 11.78AMVQHH694 pKa = 5.93VAKK697 pKa = 10.35GVRR700 pKa = 11.84ILEE703 pKa = 4.04ARR705 pKa = 11.84VARR708 pKa = 11.84VEE710 pKa = 4.57AITDD714 pKa = 4.42RR715 pKa = 11.84IMLYY719 pKa = 10.18QEE721 pKa = 4.25LDD723 pKa = 3.1CWHH726 pKa = 5.77YY727 pKa = 10.7HH728 pKa = 4.7QYY730 pKa = 11.15CVTSTRR736 pKa = 11.84AEE738 pKa = 3.88VAKK741 pKa = 10.3YY742 pKa = 10.58INWTRR747 pKa = 11.84FKK749 pKa = 11.2DD750 pKa = 3.51NCTWQQWEE758 pKa = 4.66RR759 pKa = 11.84EE760 pKa = 4.31LQGHH764 pKa = 6.37DD765 pKa = 3.84EE766 pKa = 4.32NLTMLLRR773 pKa = 11.84EE774 pKa = 4.5SARR777 pKa = 11.84MTQLAQEE784 pKa = 4.21QARR787 pKa = 11.84RR788 pKa = 11.84IPDD791 pKa = 2.85VWEE794 pKa = 4.19ALQQVFDD801 pKa = 3.76WSGWFSWLKK810 pKa = 9.6YY811 pKa = 9.94IPLIVIGILGCIIIRR826 pKa = 11.84IMICVCQPLIQVYY839 pKa = 7.94RR840 pKa = 11.84TLSTPRR846 pKa = 11.84YY847 pKa = 8.51QKK849 pKa = 9.22VTVIMEE855 pKa = 4.04TRR857 pKa = 11.84ADD859 pKa = 3.56VGGEE863 pKa = 3.74NAQDD867 pKa = 3.42FGNGFEE873 pKa = 4.1EE874 pKa = 5.02LEE876 pKa = 4.36ISRR879 pKa = 11.84EE880 pKa = 4.03PQKK883 pKa = 10.38VTIKK887 pKa = 10.54KK888 pKa = 8.57AWSNAWEE895 pKa = 4.11LWEE898 pKa = 3.92TSPWEE903 pKa = 4.05EE904 pKa = 3.25RR905 pKa = 11.84WKK907 pKa = 11.21KK908 pKa = 10.56NLLKK912 pKa = 10.42PLILPLTMGIWINGLLGEE930 pKa = 4.06RR931 pKa = 11.84HH932 pKa = 5.71KK933 pKa = 11.24NKK935 pKa = 10.06KK936 pKa = 9.39EE937 pKa = 3.97RR938 pKa = 11.84VDD940 pKa = 3.57CEE942 pKa = 3.84TWEE945 pKa = 4.58KK946 pKa = 11.38GDD948 pKa = 3.53
Molecular weight: 109.08 kDa
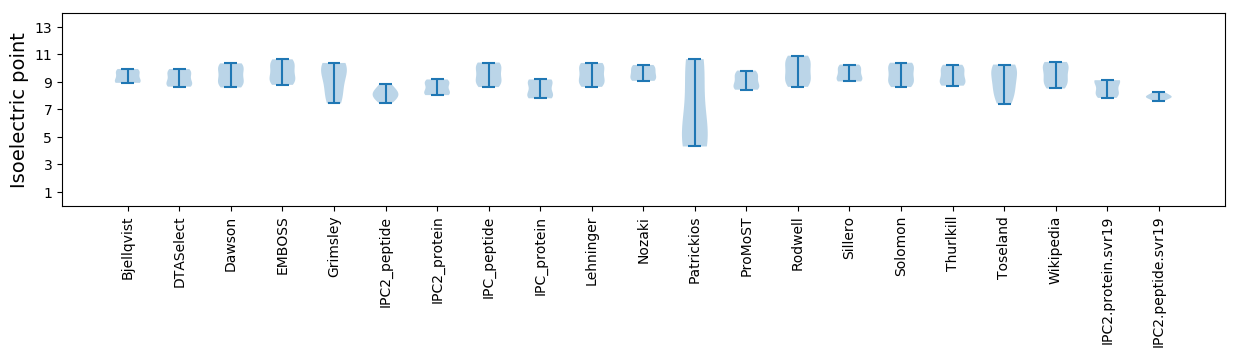
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0JIH2|C0JIH2_CAEV Env polyprotein OS=Caprine arthritis encephalitis virus Ov496 OX=621214 PE=4 SV=1
MM1 pKa = 7.63NEE3 pKa = 3.87QPPQGRR9 pKa = 11.84EE10 pKa = 3.72LHH12 pKa = 6.37PEE14 pKa = 4.3EE15 pKa = 4.35IVMSIWEE22 pKa = 4.32RR23 pKa = 11.84EE24 pKa = 3.89RR25 pKa = 11.84EE26 pKa = 4.0TWQWTSVRR34 pKa = 11.84VPNEE38 pKa = 3.13ILQRR42 pKa = 11.84WLAMLKK48 pKa = 9.89IGRR51 pKa = 11.84NRR53 pKa = 11.84GKK55 pKa = 10.4VYY57 pKa = 11.03KK58 pKa = 10.35EE59 pKa = 3.52MQKK62 pKa = 10.35WMWRR66 pKa = 11.84HH67 pKa = 5.34PKK69 pKa = 10.42APVIRR74 pKa = 11.84PCGCRR79 pKa = 11.84LCNPGWGTT87 pKa = 3.57
MM1 pKa = 7.63NEE3 pKa = 3.87QPPQGRR9 pKa = 11.84EE10 pKa = 3.72LHH12 pKa = 6.37PEE14 pKa = 4.3EE15 pKa = 4.35IVMSIWEE22 pKa = 4.32RR23 pKa = 11.84EE24 pKa = 3.89RR25 pKa = 11.84EE26 pKa = 4.0TWQWTSVRR34 pKa = 11.84VPNEE38 pKa = 3.13ILQRR42 pKa = 11.84WLAMLKK48 pKa = 9.89IGRR51 pKa = 11.84NRR53 pKa = 11.84GKK55 pKa = 10.4VYY57 pKa = 11.03KK58 pKa = 10.35EE59 pKa = 3.52MQKK62 pKa = 10.35WMWRR66 pKa = 11.84HH67 pKa = 5.34PKK69 pKa = 10.42APVIRR74 pKa = 11.84PCGCRR79 pKa = 11.84LCNPGWGTT87 pKa = 3.57
Molecular weight: 10.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2944 |
87 |
1100 |
490.7 |
56.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.74 ± 0.563 | 2.276 ± 0.584 |
3.668 ± 0.338 | 7.948 ± 0.538 |
2.106 ± 0.322 | 7.643 ± 0.438 |
2.276 ± 0.159 | 6.827 ± 1.275 |
8.016 ± 0.733 | 7.541 ± 0.286 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.057 ± 0.654 | 4.416 ± 0.616 |
4.688 ± 0.406 | 6.012 ± 0.589 |
6.42 ± 0.866 | 4.042 ± 0.315 |
5.265 ± 0.384 | 5.095 ± 0.25 |
3.906 ± 0.753 | 3.057 ± 0.421 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
