
Desulfuromusa kysingii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfuromonadales; Desulfuromonadaceae; Desulfuromusa
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
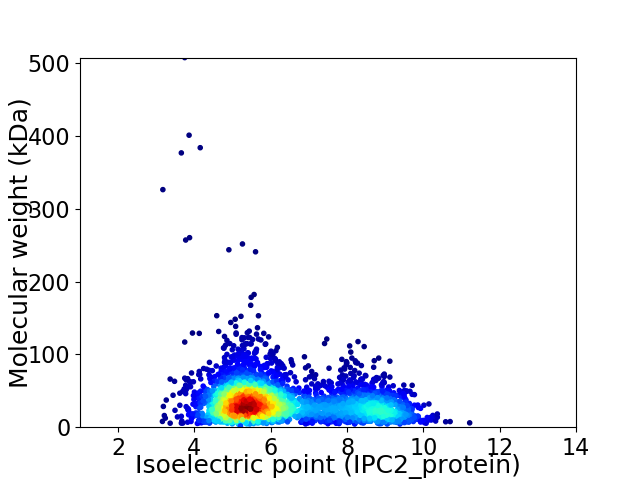
Virtual 2D-PAGE plot for 3339 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3Y870|A0A1H3Y870_9DELT Tautomerase OS=Desulfuromusa kysingii OX=37625 GN=SAMN05660420_01110 PE=3 SV=1
MM1 pKa = 7.42KK2 pKa = 9.9RR3 pKa = 11.84QIIATFILLILLVGTISTNVWAKK26 pKa = 9.5TVTLSWDD33 pKa = 3.35ASPSNVSGYY42 pKa = 10.19KK43 pKa = 9.49IYY45 pKa = 11.05YY46 pKa = 7.52GTEE49 pKa = 3.92STGSWNGSGATEE61 pKa = 4.31GDD63 pKa = 3.75SPLLVGDD70 pKa = 3.47VLTYY74 pKa = 10.6VIHH77 pKa = 7.54DD78 pKa = 4.61LPDD81 pKa = 4.62DD82 pKa = 3.77MDD84 pKa = 4.45HH85 pKa = 6.39YY86 pKa = 11.1FAVTAYY92 pKa = 10.29DD93 pKa = 3.94DD94 pKa = 3.97SDD96 pKa = 4.12NEE98 pKa = 4.36STYY101 pKa = 11.61SNIVHH106 pKa = 6.2SPVISHH112 pKa = 6.73SNTPPVLAPIGDD124 pKa = 3.83YY125 pKa = 10.77SISEE129 pKa = 4.24GATLHH134 pKa = 6.48FNLTATDD141 pKa = 3.87ADD143 pKa = 4.45GDD145 pKa = 4.28PLTYY149 pKa = 9.56TASNLPNGAGIDD161 pKa = 3.52AATGDD166 pKa = 4.47FSWTPTFDD174 pKa = 2.78QSQIYY179 pKa = 10.11AVTFSVSDD187 pKa = 3.89GEE189 pKa = 4.67SVDD192 pKa = 3.75TEE194 pKa = 4.56TINITVADD202 pKa = 3.98VALNRR207 pKa = 11.84APEE210 pKa = 4.1LSAIGNKK217 pKa = 9.44AVSEE221 pKa = 4.5GEE223 pKa = 4.04TQYY226 pKa = 11.22IPVSATDD233 pKa = 3.7PDD235 pKa = 4.15GDD237 pKa = 3.74NLVYY241 pKa = 10.71SVTNLPSGATFDD253 pKa = 3.38AANRR257 pKa = 11.84IFTWPTNFDD266 pKa = 3.09QAGIYY271 pKa = 10.53SLTFYY276 pKa = 11.24VSDD279 pKa = 3.61GTLTDD284 pKa = 3.91SEE286 pKa = 4.67QISITINNVNRR297 pKa = 11.84PPVLNSVGTQTVGEE311 pKa = 4.3GEE313 pKa = 4.01RR314 pKa = 11.84LSFTVSGVDD323 pKa = 3.34SDD325 pKa = 5.38GDD327 pKa = 3.84ALDD330 pKa = 4.68YY331 pKa = 10.97SAQNLPSGATFDD343 pKa = 3.71PLQRR347 pKa = 11.84LFSWTPSFEE356 pKa = 4.38ASTNTRR362 pKa = 11.84VYY364 pKa = 9.94PVTFTVSDD372 pKa = 4.38GSAEE376 pKa = 4.08DD377 pKa = 4.12SEE379 pKa = 5.15TITLNVTNVNRR390 pKa = 11.84APVLDD395 pKa = 4.91AIGTQVITEE404 pKa = 4.2GDD406 pKa = 3.95SVNLVVNASDD416 pKa = 4.29PDD418 pKa = 4.21NNPLTYY424 pKa = 10.22SASGLPAGAVFTAEE438 pKa = 3.83TFSFNWIPGNEE449 pKa = 4.1QSGSYY454 pKa = 7.2QVKK457 pKa = 9.24FVVSDD462 pKa = 4.02GSLSDD467 pKa = 3.89SEE469 pKa = 4.77TVTFTVNNGNEE480 pKa = 4.13PPVFSAIGAQSVEE493 pKa = 4.45EE494 pKa = 3.99NSPLSFVVVASDD506 pKa = 4.0SNGDD510 pKa = 3.72SLSYY514 pKa = 10.47SVQGLPDD521 pKa = 3.47GAVFDD526 pKa = 4.06EE527 pKa = 4.39SQRR530 pKa = 11.84RR531 pKa = 11.84FSWTPDD537 pKa = 3.12YY538 pKa = 10.29TQAGSFTVVFKK549 pKa = 11.24ASDD552 pKa = 3.67GTLSDD557 pKa = 3.98AEE559 pKa = 4.44TVEE562 pKa = 4.13ITVTNNNRR570 pKa = 11.84SPVVSGNPNGSAMATARR587 pKa = 11.84YY588 pKa = 9.89SFTPVASDD596 pKa = 3.9PDD598 pKa = 4.13GDD600 pKa = 4.08PLIFSITNKK609 pKa = 10.0PSWADD614 pKa = 3.27FSTATGEE621 pKa = 4.47LSGTPTAAQVGTSSDD636 pKa = 3.07ILISVSDD643 pKa = 4.49SISTMSLSPFSIEE656 pKa = 3.38VLAYY660 pKa = 9.54VQQDD664 pKa = 3.14SDD666 pKa = 3.67GDD668 pKa = 4.31GILDD672 pKa = 4.16HH673 pKa = 7.35LDD675 pKa = 3.84AFPDD679 pKa = 4.91DD680 pKa = 3.73NSEE683 pKa = 3.97WLDD686 pKa = 3.47TDD688 pKa = 4.14GDD690 pKa = 4.19QIGNNSDD697 pKa = 4.81LDD699 pKa = 4.15DD700 pKa = 5.84DD701 pKa = 4.61NDD703 pKa = 4.64GIADD707 pKa = 3.69VRR709 pKa = 11.84DD710 pKa = 3.94GFPLDD715 pKa = 3.65STQSGWVISATAGTGGYY732 pKa = 7.55LTPEE736 pKa = 4.45GDD738 pKa = 3.47TAVLYY743 pKa = 10.74GGSQGYY749 pKa = 8.73QLTPMSGYY757 pKa = 10.15YY758 pKa = 9.63IHH760 pKa = 7.82DD761 pKa = 4.14LLVDD765 pKa = 3.72NVSVGTVDD773 pKa = 3.5RR774 pKa = 11.84YY775 pKa = 10.07EE776 pKa = 4.1FEE778 pKa = 5.16NIGAHH783 pKa = 4.8HH784 pKa = 7.71TITAVFATIPTGLSYY799 pKa = 11.4NPISSGLIGVEE810 pKa = 3.99RR811 pKa = 11.84VDD813 pKa = 5.85GGDD816 pKa = 3.6DD817 pKa = 4.16SNNLVDD823 pKa = 5.83NKK825 pKa = 10.25PKK827 pKa = 10.71QNLDD831 pKa = 3.35YY832 pKa = 10.71QFKK835 pKa = 10.33VVLRR839 pKa = 11.84DD840 pKa = 3.68SVPADD845 pKa = 3.27QRR847 pKa = 11.84RR848 pKa = 11.84VFLILDD854 pKa = 3.33QYY856 pKa = 10.31KK857 pKa = 9.85YY858 pKa = 11.23EE859 pKa = 4.13MEE861 pKa = 4.37FDD863 pKa = 3.82SGLLSSGADD872 pKa = 3.35YY873 pKa = 11.5VLTARR878 pKa = 11.84LGPAFSHH885 pKa = 6.18SFHH888 pKa = 6.96FSAEE892 pKa = 4.13DD893 pKa = 3.23SSGNHH898 pKa = 4.67IWRR901 pKa = 11.84YY902 pKa = 7.08PASGDD907 pKa = 3.36LPGPVVSLLNGKK919 pKa = 9.0NVVGLAAKK927 pKa = 10.04INAYY931 pKa = 10.5ALTATEE937 pKa = 4.98AFNDD941 pKa = 3.64SQIYY945 pKa = 9.22RR946 pKa = 11.84WVPQSEE952 pKa = 4.19ISGHH956 pKa = 5.89FEE958 pKa = 3.99LVDD961 pKa = 3.32IGAPIATGEE970 pKa = 4.42GYY972 pKa = 9.98VLKK975 pKa = 9.54NTRR978 pKa = 11.84QSSLPNFSLYY988 pKa = 11.16GDD990 pKa = 3.33ISEE993 pKa = 4.69ATHH996 pKa = 6.47EE997 pKa = 4.46FQVHH1001 pKa = 5.9SGWNLISNPYY1011 pKa = 9.02GGNVSLADD1019 pKa = 3.27IEE1021 pKa = 4.59IRR1023 pKa = 11.84LGDD1026 pKa = 4.37ADD1028 pKa = 4.02PVPWLTAVADD1038 pKa = 3.87NLVVDD1043 pKa = 4.98VIYY1046 pKa = 10.71SYY1048 pKa = 11.5LGADD1052 pKa = 3.12WDD1054 pKa = 4.13NGNEE1058 pKa = 3.99FASAAGSKK1066 pKa = 10.17SAILVPWVGYY1076 pKa = 9.03WIYY1079 pKa = 11.41VNPTDD1084 pKa = 4.91QEE1086 pKa = 3.88ASLIILKK1093 pKa = 9.53PLQDD1097 pKa = 3.35
MM1 pKa = 7.42KK2 pKa = 9.9RR3 pKa = 11.84QIIATFILLILLVGTISTNVWAKK26 pKa = 9.5TVTLSWDD33 pKa = 3.35ASPSNVSGYY42 pKa = 10.19KK43 pKa = 9.49IYY45 pKa = 11.05YY46 pKa = 7.52GTEE49 pKa = 3.92STGSWNGSGATEE61 pKa = 4.31GDD63 pKa = 3.75SPLLVGDD70 pKa = 3.47VLTYY74 pKa = 10.6VIHH77 pKa = 7.54DD78 pKa = 4.61LPDD81 pKa = 4.62DD82 pKa = 3.77MDD84 pKa = 4.45HH85 pKa = 6.39YY86 pKa = 11.1FAVTAYY92 pKa = 10.29DD93 pKa = 3.94DD94 pKa = 3.97SDD96 pKa = 4.12NEE98 pKa = 4.36STYY101 pKa = 11.61SNIVHH106 pKa = 6.2SPVISHH112 pKa = 6.73SNTPPVLAPIGDD124 pKa = 3.83YY125 pKa = 10.77SISEE129 pKa = 4.24GATLHH134 pKa = 6.48FNLTATDD141 pKa = 3.87ADD143 pKa = 4.45GDD145 pKa = 4.28PLTYY149 pKa = 9.56TASNLPNGAGIDD161 pKa = 3.52AATGDD166 pKa = 4.47FSWTPTFDD174 pKa = 2.78QSQIYY179 pKa = 10.11AVTFSVSDD187 pKa = 3.89GEE189 pKa = 4.67SVDD192 pKa = 3.75TEE194 pKa = 4.56TINITVADD202 pKa = 3.98VALNRR207 pKa = 11.84APEE210 pKa = 4.1LSAIGNKK217 pKa = 9.44AVSEE221 pKa = 4.5GEE223 pKa = 4.04TQYY226 pKa = 11.22IPVSATDD233 pKa = 3.7PDD235 pKa = 4.15GDD237 pKa = 3.74NLVYY241 pKa = 10.71SVTNLPSGATFDD253 pKa = 3.38AANRR257 pKa = 11.84IFTWPTNFDD266 pKa = 3.09QAGIYY271 pKa = 10.53SLTFYY276 pKa = 11.24VSDD279 pKa = 3.61GTLTDD284 pKa = 3.91SEE286 pKa = 4.67QISITINNVNRR297 pKa = 11.84PPVLNSVGTQTVGEE311 pKa = 4.3GEE313 pKa = 4.01RR314 pKa = 11.84LSFTVSGVDD323 pKa = 3.34SDD325 pKa = 5.38GDD327 pKa = 3.84ALDD330 pKa = 4.68YY331 pKa = 10.97SAQNLPSGATFDD343 pKa = 3.71PLQRR347 pKa = 11.84LFSWTPSFEE356 pKa = 4.38ASTNTRR362 pKa = 11.84VYY364 pKa = 9.94PVTFTVSDD372 pKa = 4.38GSAEE376 pKa = 4.08DD377 pKa = 4.12SEE379 pKa = 5.15TITLNVTNVNRR390 pKa = 11.84APVLDD395 pKa = 4.91AIGTQVITEE404 pKa = 4.2GDD406 pKa = 3.95SVNLVVNASDD416 pKa = 4.29PDD418 pKa = 4.21NNPLTYY424 pKa = 10.22SASGLPAGAVFTAEE438 pKa = 3.83TFSFNWIPGNEE449 pKa = 4.1QSGSYY454 pKa = 7.2QVKK457 pKa = 9.24FVVSDD462 pKa = 4.02GSLSDD467 pKa = 3.89SEE469 pKa = 4.77TVTFTVNNGNEE480 pKa = 4.13PPVFSAIGAQSVEE493 pKa = 4.45EE494 pKa = 3.99NSPLSFVVVASDD506 pKa = 4.0SNGDD510 pKa = 3.72SLSYY514 pKa = 10.47SVQGLPDD521 pKa = 3.47GAVFDD526 pKa = 4.06EE527 pKa = 4.39SQRR530 pKa = 11.84RR531 pKa = 11.84FSWTPDD537 pKa = 3.12YY538 pKa = 10.29TQAGSFTVVFKK549 pKa = 11.24ASDD552 pKa = 3.67GTLSDD557 pKa = 3.98AEE559 pKa = 4.44TVEE562 pKa = 4.13ITVTNNNRR570 pKa = 11.84SPVVSGNPNGSAMATARR587 pKa = 11.84YY588 pKa = 9.89SFTPVASDD596 pKa = 3.9PDD598 pKa = 4.13GDD600 pKa = 4.08PLIFSITNKK609 pKa = 10.0PSWADD614 pKa = 3.27FSTATGEE621 pKa = 4.47LSGTPTAAQVGTSSDD636 pKa = 3.07ILISVSDD643 pKa = 4.49SISTMSLSPFSIEE656 pKa = 3.38VLAYY660 pKa = 9.54VQQDD664 pKa = 3.14SDD666 pKa = 3.67GDD668 pKa = 4.31GILDD672 pKa = 4.16HH673 pKa = 7.35LDD675 pKa = 3.84AFPDD679 pKa = 4.91DD680 pKa = 3.73NSEE683 pKa = 3.97WLDD686 pKa = 3.47TDD688 pKa = 4.14GDD690 pKa = 4.19QIGNNSDD697 pKa = 4.81LDD699 pKa = 4.15DD700 pKa = 5.84DD701 pKa = 4.61NDD703 pKa = 4.64GIADD707 pKa = 3.69VRR709 pKa = 11.84DD710 pKa = 3.94GFPLDD715 pKa = 3.65STQSGWVISATAGTGGYY732 pKa = 7.55LTPEE736 pKa = 4.45GDD738 pKa = 3.47TAVLYY743 pKa = 10.74GGSQGYY749 pKa = 8.73QLTPMSGYY757 pKa = 10.15YY758 pKa = 9.63IHH760 pKa = 7.82DD761 pKa = 4.14LLVDD765 pKa = 3.72NVSVGTVDD773 pKa = 3.5RR774 pKa = 11.84YY775 pKa = 10.07EE776 pKa = 4.1FEE778 pKa = 5.16NIGAHH783 pKa = 4.8HH784 pKa = 7.71TITAVFATIPTGLSYY799 pKa = 11.4NPISSGLIGVEE810 pKa = 3.99RR811 pKa = 11.84VDD813 pKa = 5.85GGDD816 pKa = 3.6DD817 pKa = 4.16SNNLVDD823 pKa = 5.83NKK825 pKa = 10.25PKK827 pKa = 10.71QNLDD831 pKa = 3.35YY832 pKa = 10.71QFKK835 pKa = 10.33VVLRR839 pKa = 11.84DD840 pKa = 3.68SVPADD845 pKa = 3.27QRR847 pKa = 11.84RR848 pKa = 11.84VFLILDD854 pKa = 3.33QYY856 pKa = 10.31KK857 pKa = 9.85YY858 pKa = 11.23EE859 pKa = 4.13MEE861 pKa = 4.37FDD863 pKa = 3.82SGLLSSGADD872 pKa = 3.35YY873 pKa = 11.5VLTARR878 pKa = 11.84LGPAFSHH885 pKa = 6.18SFHH888 pKa = 6.96FSAEE892 pKa = 4.13DD893 pKa = 3.23SSGNHH898 pKa = 4.67IWRR901 pKa = 11.84YY902 pKa = 7.08PASGDD907 pKa = 3.36LPGPVVSLLNGKK919 pKa = 9.0NVVGLAAKK927 pKa = 10.04INAYY931 pKa = 10.5ALTATEE937 pKa = 4.98AFNDD941 pKa = 3.64SQIYY945 pKa = 9.22RR946 pKa = 11.84WVPQSEE952 pKa = 4.19ISGHH956 pKa = 5.89FEE958 pKa = 3.99LVDD961 pKa = 3.32IGAPIATGEE970 pKa = 4.42GYY972 pKa = 9.98VLKK975 pKa = 9.54NTRR978 pKa = 11.84QSSLPNFSLYY988 pKa = 11.16GDD990 pKa = 3.33ISEE993 pKa = 4.69ATHH996 pKa = 6.47EE997 pKa = 4.46FQVHH1001 pKa = 5.9SGWNLISNPYY1011 pKa = 9.02GGNVSLADD1019 pKa = 3.27IEE1021 pKa = 4.59IRR1023 pKa = 11.84LGDD1026 pKa = 4.37ADD1028 pKa = 4.02PVPWLTAVADD1038 pKa = 3.87NLVVDD1043 pKa = 4.98VIYY1046 pKa = 10.71SYY1048 pKa = 11.5LGADD1052 pKa = 3.12WDD1054 pKa = 4.13NGNEE1058 pKa = 3.99FASAAGSKK1066 pKa = 10.17SAILVPWVGYY1076 pKa = 9.03WIYY1079 pKa = 11.41VNPTDD1084 pKa = 4.91QEE1086 pKa = 3.88ASLIILKK1093 pKa = 9.53PLQDD1097 pKa = 3.35
Molecular weight: 116.92 kDa
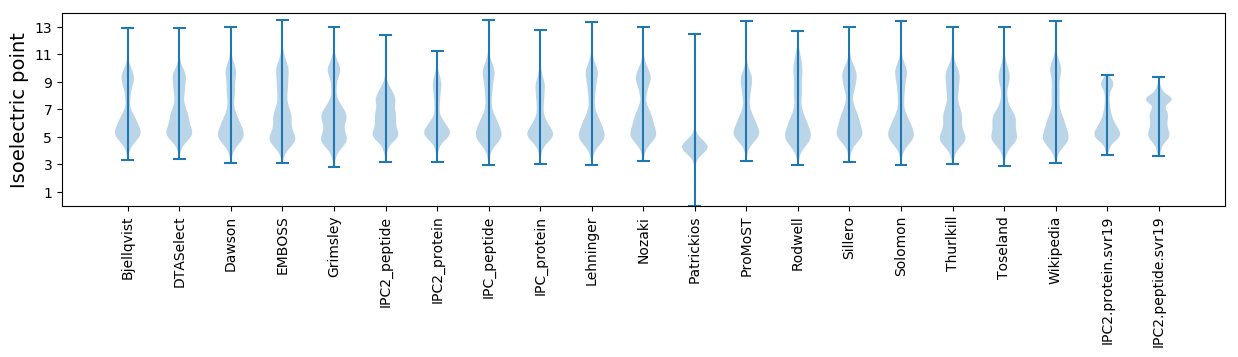
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H4AKL5|A0A1H4AKL5_9DELT ATP synthase subunit delta OS=Desulfuromusa kysingii OX=37625 GN=atpH PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPSRR10 pKa = 11.84IKK12 pKa = 10.54RR13 pKa = 11.84KK14 pKa = 8.04RR15 pKa = 11.84THH17 pKa = 6.09GFRR20 pKa = 11.84KK21 pKa = 9.94KK22 pKa = 9.27MQTKK26 pKa = 10.13SGRR29 pKa = 11.84NTINNRR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.88SLTVSIPQKK50 pKa = 10.79
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPSRR10 pKa = 11.84IKK12 pKa = 10.54RR13 pKa = 11.84KK14 pKa = 8.04RR15 pKa = 11.84THH17 pKa = 6.09GFRR20 pKa = 11.84KK21 pKa = 9.94KK22 pKa = 9.27MQTKK26 pKa = 10.13SGRR29 pKa = 11.84NTINNRR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.88SLTVSIPQKK50 pKa = 10.79
Molecular weight: 5.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1111176 |
15 |
5045 |
332.8 |
36.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.416 ± 0.047 | 1.183 ± 0.017 |
5.489 ± 0.05 | 6.237 ± 0.041 |
4.17 ± 0.035 | 7.392 ± 0.088 |
2.063 ± 0.023 | 6.751 ± 0.032 |
5.115 ± 0.044 | 11.004 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.024 | 3.784 ± 0.032 |
4.1 ± 0.031 | 4.461 ± 0.043 |
4.991 ± 0.041 | 6.268 ± 0.041 |
5.32 ± 0.036 | 6.879 ± 0.037 |
1.009 ± 0.016 | 2.898 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
