
Changjiang tombus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.7
Get precalculated fractions of proteins
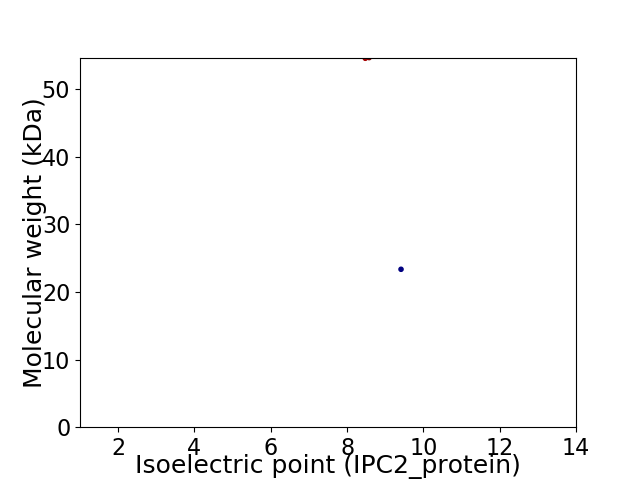
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
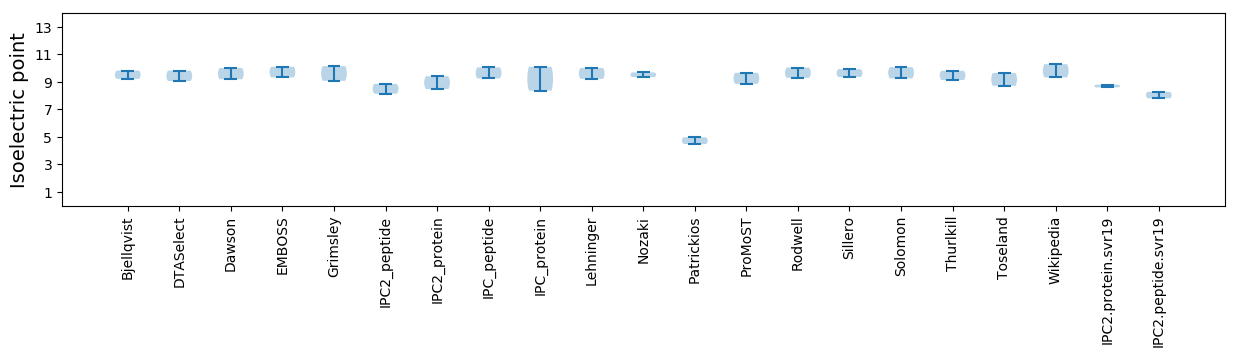
Protein with the lowest isoelectric point:
>tr|A0A1L3KFP0|A0A1L3KFP0_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 1 OX=1922802 PE=4 SV=1
MM1 pKa = 7.69GEE3 pKa = 4.4FVCDD7 pKa = 5.47DD8 pKa = 3.71IMTTHH13 pKa = 6.42NNSIANLIRR22 pKa = 11.84GVGEE26 pKa = 3.52RR27 pKa = 11.84VLFTDD32 pKa = 4.64RR33 pKa = 11.84QLTPCIKK40 pKa = 9.61PLVGIFSKK48 pKa = 10.76RR49 pKa = 11.84CASFSRR55 pKa = 11.84EE56 pKa = 3.59VARR59 pKa = 11.84CVGRR63 pKa = 11.84QSPVSRR69 pKa = 11.84QSFVDD74 pKa = 3.67YY75 pKa = 10.84YY76 pKa = 10.35KK77 pKa = 10.77GRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84TIYY84 pKa = 9.8QQAADD89 pKa = 4.16GLVLKK94 pKa = 9.47PIRR97 pKa = 11.84PRR99 pKa = 11.84DD100 pKa = 3.56AYY102 pKa = 11.23LSTFVKK108 pKa = 10.54AEE110 pKa = 4.34KK111 pKa = 10.19INLSNKK117 pKa = 8.83PDD119 pKa = 4.04PAPRR123 pKa = 11.84VIQPRR128 pKa = 11.84NPRR131 pKa = 11.84YY132 pKa = 9.44NVEE135 pKa = 4.17LGRR138 pKa = 11.84FLLPLEE144 pKa = 4.57HH145 pKa = 6.72KK146 pKa = 10.79VYY148 pKa = 10.89DD149 pKa = 4.97AIDD152 pKa = 3.68EE153 pKa = 4.59VFGSPTIMSKK163 pKa = 10.97YY164 pKa = 10.13NSVDD168 pKa = 2.95QAAVLRR174 pKa = 11.84EE175 pKa = 3.74KK176 pKa = 9.71WDD178 pKa = 3.38KK179 pKa = 10.63FRR181 pKa = 11.84EE182 pKa = 4.29PVCVGLDD189 pKa = 3.18ASRR192 pKa = 11.84FDD194 pKa = 3.47QHH196 pKa = 8.35VSEE199 pKa = 4.21QALKK203 pKa = 10.63FEE205 pKa = 4.79HH206 pKa = 7.12DD207 pKa = 4.57FYY209 pKa = 11.85RR210 pKa = 11.84MVFGPRR216 pKa = 11.84KK217 pKa = 9.3DD218 pKa = 4.04LSMLLSWQLTNKK230 pKa = 9.64GFARR234 pKa = 11.84ATDD237 pKa = 3.93GSFSYY242 pKa = 10.15IRR244 pKa = 11.84KK245 pKa = 8.1GSRR248 pKa = 11.84MSGDD252 pKa = 3.39MNTSLGNKK260 pKa = 9.47FLMCAMGKK268 pKa = 8.24TYY270 pKa = 10.86LDD272 pKa = 3.34QLQIKK277 pKa = 9.93YY278 pKa = 10.16EE279 pKa = 4.0FANNGDD285 pKa = 3.68DD286 pKa = 3.3CLVFIEE292 pKa = 4.79RR293 pKa = 11.84SNLKK297 pKa = 10.09KK298 pKa = 10.86LSGLQTYY305 pKa = 9.45FRR307 pKa = 11.84GFGFKK312 pKa = 9.81IVTEE316 pKa = 4.1KK317 pKa = 10.74PVFEE321 pKa = 4.46FEE323 pKa = 4.51HH324 pKa = 8.62VEE326 pKa = 4.36FCQCRR331 pKa = 11.84PLFCNNIWRR340 pKa = 11.84MVRR343 pKa = 11.84NVKK346 pKa = 8.7TCLLKK351 pKa = 10.93DD352 pKa = 3.57VTSVNLGHH360 pKa = 7.74DD361 pKa = 3.83VTQYY365 pKa = 10.82RR366 pKa = 11.84AWLADD371 pKa = 3.06IAGCGLSFAADD382 pKa = 3.65VPILGAYY389 pKa = 9.86YY390 pKa = 10.97RR391 pKa = 11.84MLQRR395 pKa = 11.84FGQAGRR401 pKa = 11.84YY402 pKa = 9.2HH403 pKa = 7.22GDD405 pKa = 2.98DD406 pKa = 4.18SMFNCYY412 pKa = 7.93RR413 pKa = 11.84TLSKK417 pKa = 10.76NSRR420 pKa = 11.84ISSTVPDD427 pKa = 3.6ALGRR431 pKa = 11.84YY432 pKa = 7.8SFWLQTGIHH441 pKa = 7.29PDD443 pKa = 3.29GQQAIEE449 pKa = 4.34NYY451 pKa = 9.84FEE453 pKa = 3.76EE454 pKa = 5.64AIWGGDD460 pKa = 2.94KK461 pKa = 10.66RR462 pKa = 11.84QLINNIHH469 pKa = 6.49YY470 pKa = 10.04ILKK473 pKa = 10.54NGSS476 pKa = 3.07
MM1 pKa = 7.69GEE3 pKa = 4.4FVCDD7 pKa = 5.47DD8 pKa = 3.71IMTTHH13 pKa = 6.42NNSIANLIRR22 pKa = 11.84GVGEE26 pKa = 3.52RR27 pKa = 11.84VLFTDD32 pKa = 4.64RR33 pKa = 11.84QLTPCIKK40 pKa = 9.61PLVGIFSKK48 pKa = 10.76RR49 pKa = 11.84CASFSRR55 pKa = 11.84EE56 pKa = 3.59VARR59 pKa = 11.84CVGRR63 pKa = 11.84QSPVSRR69 pKa = 11.84QSFVDD74 pKa = 3.67YY75 pKa = 10.84YY76 pKa = 10.35KK77 pKa = 10.77GRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84TIYY84 pKa = 9.8QQAADD89 pKa = 4.16GLVLKK94 pKa = 9.47PIRR97 pKa = 11.84PRR99 pKa = 11.84DD100 pKa = 3.56AYY102 pKa = 11.23LSTFVKK108 pKa = 10.54AEE110 pKa = 4.34KK111 pKa = 10.19INLSNKK117 pKa = 8.83PDD119 pKa = 4.04PAPRR123 pKa = 11.84VIQPRR128 pKa = 11.84NPRR131 pKa = 11.84YY132 pKa = 9.44NVEE135 pKa = 4.17LGRR138 pKa = 11.84FLLPLEE144 pKa = 4.57HH145 pKa = 6.72KK146 pKa = 10.79VYY148 pKa = 10.89DD149 pKa = 4.97AIDD152 pKa = 3.68EE153 pKa = 4.59VFGSPTIMSKK163 pKa = 10.97YY164 pKa = 10.13NSVDD168 pKa = 2.95QAAVLRR174 pKa = 11.84EE175 pKa = 3.74KK176 pKa = 9.71WDD178 pKa = 3.38KK179 pKa = 10.63FRR181 pKa = 11.84EE182 pKa = 4.29PVCVGLDD189 pKa = 3.18ASRR192 pKa = 11.84FDD194 pKa = 3.47QHH196 pKa = 8.35VSEE199 pKa = 4.21QALKK203 pKa = 10.63FEE205 pKa = 4.79HH206 pKa = 7.12DD207 pKa = 4.57FYY209 pKa = 11.85RR210 pKa = 11.84MVFGPRR216 pKa = 11.84KK217 pKa = 9.3DD218 pKa = 4.04LSMLLSWQLTNKK230 pKa = 9.64GFARR234 pKa = 11.84ATDD237 pKa = 3.93GSFSYY242 pKa = 10.15IRR244 pKa = 11.84KK245 pKa = 8.1GSRR248 pKa = 11.84MSGDD252 pKa = 3.39MNTSLGNKK260 pKa = 9.47FLMCAMGKK268 pKa = 8.24TYY270 pKa = 10.86LDD272 pKa = 3.34QLQIKK277 pKa = 9.93YY278 pKa = 10.16EE279 pKa = 4.0FANNGDD285 pKa = 3.68DD286 pKa = 3.3CLVFIEE292 pKa = 4.79RR293 pKa = 11.84SNLKK297 pKa = 10.09KK298 pKa = 10.86LSGLQTYY305 pKa = 9.45FRR307 pKa = 11.84GFGFKK312 pKa = 9.81IVTEE316 pKa = 4.1KK317 pKa = 10.74PVFEE321 pKa = 4.46FEE323 pKa = 4.51HH324 pKa = 8.62VEE326 pKa = 4.36FCQCRR331 pKa = 11.84PLFCNNIWRR340 pKa = 11.84MVRR343 pKa = 11.84NVKK346 pKa = 8.7TCLLKK351 pKa = 10.93DD352 pKa = 3.57VTSVNLGHH360 pKa = 7.74DD361 pKa = 3.83VTQYY365 pKa = 10.82RR366 pKa = 11.84AWLADD371 pKa = 3.06IAGCGLSFAADD382 pKa = 3.65VPILGAYY389 pKa = 9.86YY390 pKa = 10.97RR391 pKa = 11.84MLQRR395 pKa = 11.84FGQAGRR401 pKa = 11.84YY402 pKa = 9.2HH403 pKa = 7.22GDD405 pKa = 2.98DD406 pKa = 4.18SMFNCYY412 pKa = 7.93RR413 pKa = 11.84TLSKK417 pKa = 10.76NSRR420 pKa = 11.84ISSTVPDD427 pKa = 3.6ALGRR431 pKa = 11.84YY432 pKa = 7.8SFWLQTGIHH441 pKa = 7.29PDD443 pKa = 3.29GQQAIEE449 pKa = 4.34NYY451 pKa = 9.84FEE453 pKa = 3.76EE454 pKa = 5.64AIWGGDD460 pKa = 2.94KK461 pKa = 10.66RR462 pKa = 11.84QLINNIHH469 pKa = 6.49YY470 pKa = 10.04ILKK473 pKa = 10.54NGSS476 pKa = 3.07
Molecular weight: 54.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFP0|A0A1L3KFP0_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 1 OX=1922802 PE=4 SV=1
MM1 pKa = 7.2ARR3 pKa = 11.84KK4 pKa = 8.28RR5 pKa = 11.84TRR7 pKa = 11.84SSQSSDD13 pKa = 2.83EE14 pKa = 4.28SGSLISRR21 pKa = 11.84NRR23 pKa = 11.84QPPRR27 pKa = 11.84PRR29 pKa = 11.84VRR31 pKa = 11.84GSLGTNTILRR41 pKa = 11.84GVEE44 pKa = 3.56IGGSLVTDD52 pKa = 4.38ANGSAAGVYY61 pKa = 9.59PLIAGSLTGLTNSPINNIAKK81 pKa = 9.54YY82 pKa = 9.22YY83 pKa = 10.71NSFVYY88 pKa = 10.08QSAVMHH94 pKa = 6.39YY95 pKa = 9.24IPAVGLTTPGQVTAVFINNTEE116 pKa = 3.78WMSYY120 pKa = 11.0ALDD123 pKa = 3.47GTRR126 pKa = 11.84TVTEE130 pKa = 4.78LGTLALSQSNAATHH144 pKa = 6.05AVWHH148 pKa = 5.74EE149 pKa = 3.58FSYY152 pKa = 11.53AMNLPSRR159 pKa = 11.84RR160 pKa = 11.84KK161 pKa = 9.69RR162 pKa = 11.84FDD164 pKa = 3.42VNSTLPTATVDD175 pKa = 3.56VVEE178 pKa = 5.12RR179 pKa = 11.84DD180 pKa = 3.76CQGVFILFVSGAPANTTISVPRR202 pKa = 11.84RR203 pKa = 11.84SVTLQVEE210 pKa = 4.68GMSGFFPP217 pKa = 4.87
MM1 pKa = 7.2ARR3 pKa = 11.84KK4 pKa = 8.28RR5 pKa = 11.84TRR7 pKa = 11.84SSQSSDD13 pKa = 2.83EE14 pKa = 4.28SGSLISRR21 pKa = 11.84NRR23 pKa = 11.84QPPRR27 pKa = 11.84PRR29 pKa = 11.84VRR31 pKa = 11.84GSLGTNTILRR41 pKa = 11.84GVEE44 pKa = 3.56IGGSLVTDD52 pKa = 4.38ANGSAAGVYY61 pKa = 9.59PLIAGSLTGLTNSPINNIAKK81 pKa = 9.54YY82 pKa = 9.22YY83 pKa = 10.71NSFVYY88 pKa = 10.08QSAVMHH94 pKa = 6.39YY95 pKa = 9.24IPAVGLTTPGQVTAVFINNTEE116 pKa = 3.78WMSYY120 pKa = 11.0ALDD123 pKa = 3.47GTRR126 pKa = 11.84TVTEE130 pKa = 4.78LGTLALSQSNAATHH144 pKa = 6.05AVWHH148 pKa = 5.74EE149 pKa = 3.58FSYY152 pKa = 11.53AMNLPSRR159 pKa = 11.84RR160 pKa = 11.84KK161 pKa = 9.69RR162 pKa = 11.84FDD164 pKa = 3.42VNSTLPTATVDD175 pKa = 3.56VVEE178 pKa = 5.12RR179 pKa = 11.84DD180 pKa = 3.76CQGVFILFVSGAPANTTISVPRR202 pKa = 11.84RR203 pKa = 11.84SVTLQVEE210 pKa = 4.68GMSGFFPP217 pKa = 4.87
Molecular weight: 23.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
693 |
217 |
476 |
346.5 |
38.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.494 ± 0.945 | 2.02 ± 0.818 |
5.051 ± 1.199 | 3.896 ± 0.352 |
5.483 ± 0.943 | 7.504 ± 0.415 |
1.732 ± 0.183 | 5.051 ± 0.232 |
4.329 ± 1.546 | 8.081 ± 0.371 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.453 ± 0.078 | 5.339 ± 0.342 |
4.473 ± 0.555 | 4.04 ± 0.428 |
7.792 ± 0.22 | 8.081 ± 1.563 |
5.772 ± 2.05 | 7.359 ± 0.975 |
1.154 ± 0.122 | 3.896 ± 0.352 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
