
Pacific flying fox faeces associated circular DNA virus-1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.74
Get precalculated fractions of proteins
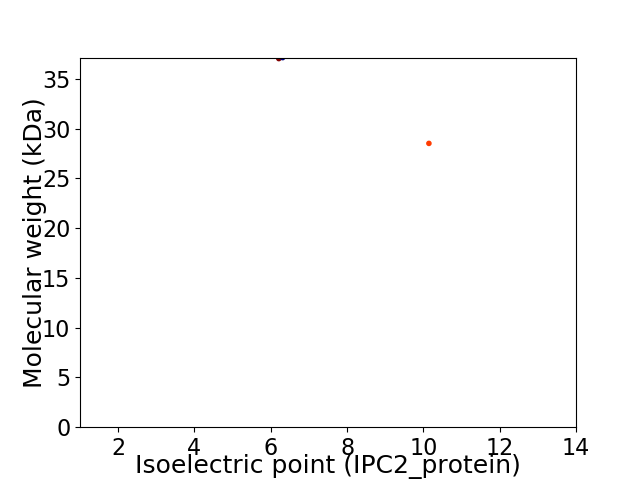
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
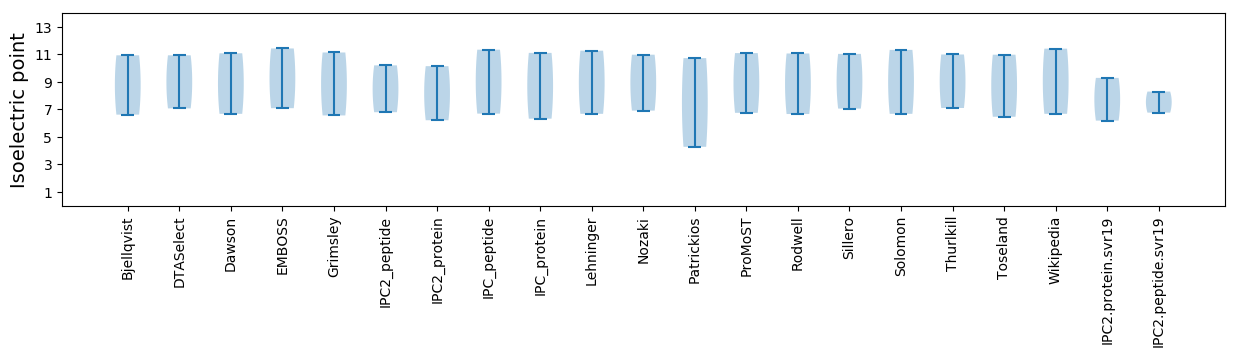
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A140CTT7|A0A140CTT7_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated circular DNA virus-1 OX=1796004 PE=4 SV=1
MM1 pKa = 7.21QLSRR5 pKa = 11.84WCFTNFNIPQDD16 pKa = 3.8LAAGCDD22 pKa = 3.65APLTLPSEE30 pKa = 4.25RR31 pKa = 11.84LKK33 pKa = 10.4YY34 pKa = 10.56ACYY37 pKa = 10.12QIEE40 pKa = 4.22KK41 pKa = 10.5CPDD44 pKa = 3.02TGRR47 pKa = 11.84LHH49 pKa = 6.12AQGYY53 pKa = 7.54LHH55 pKa = 7.26FKK57 pKa = 10.65RR58 pKa = 11.84SIKK61 pKa = 10.74LGGLKK66 pKa = 10.12KK67 pKa = 10.87LPGFEE72 pKa = 4.06AAHH75 pKa = 6.14FEE77 pKa = 4.21GAKK80 pKa = 10.31ADD82 pKa = 3.75LQANRR87 pKa = 11.84NYY89 pKa = 8.56CTKK92 pKa = 10.13EE93 pKa = 3.92DD94 pKa = 3.78SRR96 pKa = 11.84VAGPFEE102 pKa = 4.49IGSTEE107 pKa = 3.84GLGPGARR114 pKa = 11.84SDD116 pKa = 3.6LLAVKK121 pKa = 10.19RR122 pKa = 11.84RR123 pKa = 11.84LDD125 pKa = 3.51EE126 pKa = 4.57GVDD129 pKa = 3.73EE130 pKa = 5.1VGLADD135 pKa = 3.69EE136 pKa = 6.27FFGEE140 pKa = 4.61WVKK143 pKa = 10.83HH144 pKa = 4.28RR145 pKa = 11.84ASFNVYY151 pKa = 10.02RR152 pKa = 11.84GLKK155 pKa = 9.36VPARR159 pKa = 11.84GKK161 pKa = 9.0DD162 pKa = 3.41TQTPLYY168 pKa = 9.25VCVGASGLGKK178 pKa = 9.03STFVYY183 pKa = 10.64DD184 pKa = 3.69EE185 pKa = 4.4TEE187 pKa = 3.81GTVFQKK193 pKa = 11.27ALTAKK198 pKa = 8.91WWDD201 pKa = 3.86GFNGTQPVLFDD212 pKa = 3.93DD213 pKa = 4.35FRR215 pKa = 11.84GAFQFTEE222 pKa = 4.31LLRR225 pKa = 11.84LANIGAYY232 pKa = 7.81QVEE235 pKa = 4.69VKK237 pKa = 10.65GGSIQFNPKK246 pKa = 9.69EE247 pKa = 3.61IWITSNLRR255 pKa = 11.84PDD257 pKa = 2.87QWYY260 pKa = 10.02NPSTIKK266 pKa = 10.75DD267 pKa = 3.34ITPLTRR273 pKa = 11.84RR274 pKa = 11.84ISKK277 pKa = 9.8IYY279 pKa = 10.14HH280 pKa = 4.57FTAFKK285 pKa = 10.49EE286 pKa = 3.93MRR288 pKa = 11.84LFQSTEE294 pKa = 3.87NEE296 pKa = 4.58SAWDD300 pKa = 3.39QYY302 pKa = 11.53SRR304 pKa = 11.84SEE306 pKa = 4.14EE307 pKa = 3.84FSTLNFNGRR316 pKa = 11.84HH317 pKa = 5.43GEE319 pKa = 4.01ATSDD323 pKa = 4.01LEE325 pKa = 4.74LVYY328 pKa = 11.07
MM1 pKa = 7.21QLSRR5 pKa = 11.84WCFTNFNIPQDD16 pKa = 3.8LAAGCDD22 pKa = 3.65APLTLPSEE30 pKa = 4.25RR31 pKa = 11.84LKK33 pKa = 10.4YY34 pKa = 10.56ACYY37 pKa = 10.12QIEE40 pKa = 4.22KK41 pKa = 10.5CPDD44 pKa = 3.02TGRR47 pKa = 11.84LHH49 pKa = 6.12AQGYY53 pKa = 7.54LHH55 pKa = 7.26FKK57 pKa = 10.65RR58 pKa = 11.84SIKK61 pKa = 10.74LGGLKK66 pKa = 10.12KK67 pKa = 10.87LPGFEE72 pKa = 4.06AAHH75 pKa = 6.14FEE77 pKa = 4.21GAKK80 pKa = 10.31ADD82 pKa = 3.75LQANRR87 pKa = 11.84NYY89 pKa = 8.56CTKK92 pKa = 10.13EE93 pKa = 3.92DD94 pKa = 3.78SRR96 pKa = 11.84VAGPFEE102 pKa = 4.49IGSTEE107 pKa = 3.84GLGPGARR114 pKa = 11.84SDD116 pKa = 3.6LLAVKK121 pKa = 10.19RR122 pKa = 11.84RR123 pKa = 11.84LDD125 pKa = 3.51EE126 pKa = 4.57GVDD129 pKa = 3.73EE130 pKa = 5.1VGLADD135 pKa = 3.69EE136 pKa = 6.27FFGEE140 pKa = 4.61WVKK143 pKa = 10.83HH144 pKa = 4.28RR145 pKa = 11.84ASFNVYY151 pKa = 10.02RR152 pKa = 11.84GLKK155 pKa = 9.36VPARR159 pKa = 11.84GKK161 pKa = 9.0DD162 pKa = 3.41TQTPLYY168 pKa = 9.25VCVGASGLGKK178 pKa = 9.03STFVYY183 pKa = 10.64DD184 pKa = 3.69EE185 pKa = 4.4TEE187 pKa = 3.81GTVFQKK193 pKa = 11.27ALTAKK198 pKa = 8.91WWDD201 pKa = 3.86GFNGTQPVLFDD212 pKa = 3.93DD213 pKa = 4.35FRR215 pKa = 11.84GAFQFTEE222 pKa = 4.31LLRR225 pKa = 11.84LANIGAYY232 pKa = 7.81QVEE235 pKa = 4.69VKK237 pKa = 10.65GGSIQFNPKK246 pKa = 9.69EE247 pKa = 3.61IWITSNLRR255 pKa = 11.84PDD257 pKa = 2.87QWYY260 pKa = 10.02NPSTIKK266 pKa = 10.75DD267 pKa = 3.34ITPLTRR273 pKa = 11.84RR274 pKa = 11.84ISKK277 pKa = 9.8IYY279 pKa = 10.14HH280 pKa = 4.57FTAFKK285 pKa = 10.49EE286 pKa = 3.93MRR288 pKa = 11.84LFQSTEE294 pKa = 3.87NEE296 pKa = 4.58SAWDD300 pKa = 3.39QYY302 pKa = 11.53SRR304 pKa = 11.84SEE306 pKa = 4.14EE307 pKa = 3.84FSTLNFNGRR316 pKa = 11.84HH317 pKa = 5.43GEE319 pKa = 4.01ATSDD323 pKa = 4.01LEE325 pKa = 4.74LVYY328 pKa = 11.07
Molecular weight: 37.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTT7|A0A140CTT7_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated circular DNA virus-1 OX=1796004 PE=4 SV=1
MM1 pKa = 7.42PWRR4 pKa = 11.84RR5 pKa = 11.84SWKK8 pKa = 9.47RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84STYY14 pKa = 9.7RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TTRR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 9.13GRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84PWRR29 pKa = 11.84RR30 pKa = 11.84TRR32 pKa = 11.84TRR34 pKa = 11.84SMMSRR39 pKa = 11.84SRR41 pKa = 11.84NLRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GTRR50 pKa = 11.84VFTRR54 pKa = 11.84TDD56 pKa = 2.57KK57 pKa = 10.94HH58 pKa = 6.71VYY60 pKa = 7.78STVYY64 pKa = 9.0TQGNSYY70 pKa = 8.56TAGSFAPKK78 pKa = 10.16LADD81 pKa = 3.37APGYY85 pKa = 9.58EE86 pKa = 4.45NLYY89 pKa = 10.19QLYY92 pKa = 9.78DD93 pKa = 3.12QARR96 pKa = 11.84IVKK99 pKa = 9.02FSIRR103 pKa = 11.84IIPSLKK109 pKa = 8.99SQVMVPTQQLPGEE122 pKa = 4.29YY123 pKa = 9.84SSIVQQVYY131 pKa = 9.85VYY133 pKa = 11.07NATNWEE139 pKa = 4.2GTPNDD144 pKa = 3.76PNVFLEE150 pKa = 4.07QSGCRR155 pKa = 11.84SFSPYY160 pKa = 9.03KK161 pKa = 10.22TILARR166 pKa = 11.84RR167 pKa = 11.84AARR170 pKa = 11.84TLEE173 pKa = 3.83KK174 pKa = 10.42STFQNSEE181 pKa = 3.96YY182 pKa = 10.1DD183 pKa = 3.52TLEE186 pKa = 3.98QNHH189 pKa = 5.38VRR191 pKa = 11.84MHH193 pKa = 6.1PWIDD197 pKa = 2.88VDD199 pKa = 4.0YY200 pKa = 11.53GGGALNCLPIVVGFPPNQNSSPSVSQSWKK229 pKa = 9.45VEE231 pKa = 3.6ITFTHH236 pKa = 4.55QWKK239 pKa = 10.63SKK241 pKa = 9.97LL242 pKa = 3.64
MM1 pKa = 7.42PWRR4 pKa = 11.84RR5 pKa = 11.84SWKK8 pKa = 9.47RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84STYY14 pKa = 9.7RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TTRR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 9.13GRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84PWRR29 pKa = 11.84RR30 pKa = 11.84TRR32 pKa = 11.84TRR34 pKa = 11.84SMMSRR39 pKa = 11.84SRR41 pKa = 11.84NLRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GTRR50 pKa = 11.84VFTRR54 pKa = 11.84TDD56 pKa = 2.57KK57 pKa = 10.94HH58 pKa = 6.71VYY60 pKa = 7.78STVYY64 pKa = 9.0TQGNSYY70 pKa = 8.56TAGSFAPKK78 pKa = 10.16LADD81 pKa = 3.37APGYY85 pKa = 9.58EE86 pKa = 4.45NLYY89 pKa = 10.19QLYY92 pKa = 9.78DD93 pKa = 3.12QARR96 pKa = 11.84IVKK99 pKa = 9.02FSIRR103 pKa = 11.84IIPSLKK109 pKa = 8.99SQVMVPTQQLPGEE122 pKa = 4.29YY123 pKa = 9.84SSIVQQVYY131 pKa = 9.85VYY133 pKa = 11.07NATNWEE139 pKa = 4.2GTPNDD144 pKa = 3.76PNVFLEE150 pKa = 4.07QSGCRR155 pKa = 11.84SFSPYY160 pKa = 9.03KK161 pKa = 10.22TILARR166 pKa = 11.84RR167 pKa = 11.84AARR170 pKa = 11.84TLEE173 pKa = 3.83KK174 pKa = 10.42STFQNSEE181 pKa = 3.96YY182 pKa = 10.1DD183 pKa = 3.52TLEE186 pKa = 3.98QNHH189 pKa = 5.38VRR191 pKa = 11.84MHH193 pKa = 6.1PWIDD197 pKa = 2.88VDD199 pKa = 4.0YY200 pKa = 11.53GGGALNCLPIVVGFPPNQNSSPSVSQSWKK229 pKa = 9.45VEE231 pKa = 3.6ITFTHH236 pKa = 4.55QWKK239 pKa = 10.63SKK241 pKa = 9.97LL242 pKa = 3.64
Molecular weight: 28.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
570 |
242 |
328 |
285.0 |
32.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.14 ± 1.283 | 1.404 ± 0.369 |
4.386 ± 0.954 | 5.439 ± 1.362 |
5.263 ± 1.25 | 7.193 ± 1.427 |
1.754 ± 0.065 | 3.684 ± 0.022 |
5.263 ± 0.722 | 7.544 ± 1.387 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.228 ± 0.535 | 4.386 ± 0.366 |
5.088 ± 0.709 | 4.912 ± 0.557 |
9.123 ± 2.619 | 7.544 ± 1.516 |
7.193 ± 0.684 | 5.439 ± 0.749 |
2.456 ± 0.279 | 4.561 ± 0.782 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
