
Pelargonium zonate spot virus (isolate Tomato/Italy/1982) (PZSV) (Pelargonium zonate spot virus (isolate Tomato))
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Anulavirus; Pelargonium zonate spot virus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
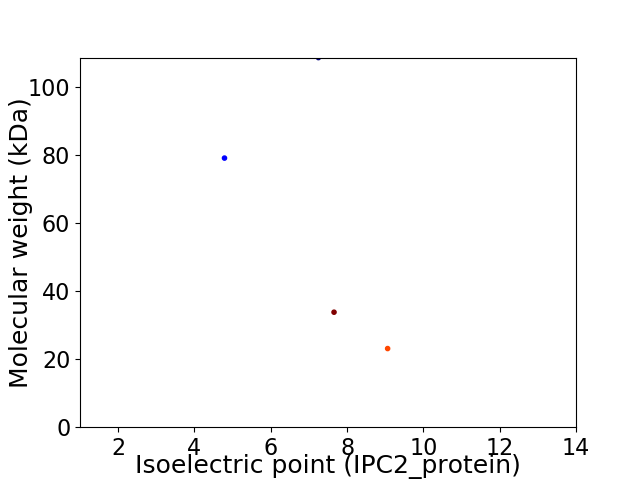
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9DUT3|1A_PZSVT Replication protein 1a OS=Pelargonium zonate spot virus (isolate Tomato/Italy/1982) OX=650488 GN=ORF1a PE=3 SV=1
MM1 pKa = 7.82AAFTFEE7 pKa = 4.28NFLSGAYY14 pKa = 8.81TGLPIDD20 pKa = 3.93KK21 pKa = 10.11FRR23 pKa = 11.84ALGLNTEE30 pKa = 5.06DD31 pKa = 4.17YY32 pKa = 11.27DD33 pKa = 3.88EE34 pKa = 4.7QRR36 pKa = 11.84WEE38 pKa = 4.05MLVKK42 pKa = 10.6CVDD45 pKa = 3.4SGLMQFSCSRR55 pKa = 11.84DD56 pKa = 3.31EE57 pKa = 4.96ALVLLWNEE65 pKa = 3.97EE66 pKa = 4.25EE67 pKa = 4.89LPKK70 pKa = 10.92DD71 pKa = 4.08DD72 pKa = 5.47DD73 pKa = 4.96DD74 pKa = 4.04EE75 pKa = 4.76VPYY78 pKa = 10.13EE79 pKa = 4.42VPCWTPDD86 pKa = 2.94TDD88 pKa = 3.41ATVIDD93 pKa = 5.79DD94 pKa = 3.71VSEE97 pKa = 3.82WLAEE101 pKa = 4.1KK102 pKa = 10.21TSVRR106 pKa = 11.84DD107 pKa = 3.64TVVVCSDD114 pKa = 3.28YY115 pKa = 11.55DD116 pKa = 3.74AVSEE120 pKa = 4.47TPVEE124 pKa = 4.22VLSVEE129 pKa = 4.14LEE131 pKa = 3.97EE132 pKa = 6.24DD133 pKa = 3.78SEE135 pKa = 4.55EE136 pKa = 3.97DD137 pKa = 3.64TIADD141 pKa = 3.34VCLDD145 pKa = 3.28ARR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 9.8SFRR152 pKa = 11.84DD153 pKa = 3.1YY154 pKa = 9.61FTIVEE159 pKa = 4.22EE160 pKa = 4.36EE161 pKa = 4.18FVEE164 pKa = 4.07EE165 pKa = 5.24DD166 pKa = 3.92PLISLNDD173 pKa = 3.66GNVCPVRR180 pKa = 11.84THH182 pKa = 6.17EE183 pKa = 4.33VTSLEE188 pKa = 4.33KK189 pKa = 10.34PVMLDD194 pKa = 2.65VGRR197 pKa = 11.84RR198 pKa = 11.84CDD200 pKa = 3.51RR201 pKa = 11.84VNLEE205 pKa = 4.18SLQGAINMNLPSHH218 pKa = 7.03AYY220 pKa = 10.24FDD222 pKa = 4.69DD223 pKa = 3.38TWHH226 pKa = 6.17QYY228 pKa = 8.96FVEE231 pKa = 4.81GSKK234 pKa = 11.0LDD236 pKa = 3.47VDD238 pKa = 4.86FDD240 pKa = 4.62NIRR243 pKa = 11.84LRR245 pKa = 11.84QSEE248 pKa = 4.72VFCDD252 pKa = 3.68RR253 pKa = 11.84DD254 pKa = 3.14LDD256 pKa = 4.15RR257 pKa = 11.84YY258 pKa = 8.31YY259 pKa = 10.73QPEE262 pKa = 4.46LFAGASSRR270 pKa = 11.84RR271 pKa = 11.84IGTQKK276 pKa = 10.21EE277 pKa = 3.7ALVAIRR283 pKa = 11.84KK284 pKa = 9.45RR285 pKa = 11.84NADD288 pKa = 3.54VPEE291 pKa = 4.47LADD294 pKa = 3.59SVDD297 pKa = 3.52VEE299 pKa = 4.29RR300 pKa = 11.84LSEE303 pKa = 4.23SVAKK307 pKa = 10.37KK308 pKa = 9.94FLSSYY313 pKa = 10.69VCDD316 pKa = 4.51LKK318 pKa = 11.16PVVGVMEE325 pKa = 4.54KK326 pKa = 9.75MRR328 pKa = 11.84AYY330 pKa = 8.88HH331 pKa = 6.01QKK333 pKa = 9.6WGDD336 pKa = 3.54KK337 pKa = 9.49IDD339 pKa = 3.6PMFLLKK345 pKa = 10.27EE346 pKa = 4.29HH347 pKa = 6.39NLQRR351 pKa = 11.84YY352 pKa = 5.82EE353 pKa = 4.23HH354 pKa = 6.57MIKK357 pKa = 9.59TDD359 pKa = 3.37VKK361 pKa = 8.52PTVAHH366 pKa = 5.81SMHH369 pKa = 6.59VEE371 pKa = 3.75RR372 pKa = 11.84AIPATITFHH381 pKa = 6.56GKK383 pKa = 9.69SICAGFSPSFTALFDD398 pKa = 3.86EE399 pKa = 5.18FQKK402 pKa = 11.26SLDD405 pKa = 3.69EE406 pKa = 4.08RR407 pKa = 11.84VVIPSGPISTIEE419 pKa = 3.64MDD421 pKa = 3.65FDD423 pKa = 3.66IRR425 pKa = 11.84NKK427 pKa = 10.67YY428 pKa = 8.51YY429 pKa = 11.19LEE431 pKa = 4.25VDD433 pKa = 3.8LSKK436 pKa = 10.71FDD438 pKa = 3.62KK439 pKa = 11.14SQGLLHH445 pKa = 7.4LEE447 pKa = 4.19FQRR450 pKa = 11.84KK451 pKa = 7.97ILCKK455 pKa = 9.97IGLPAHH461 pKa = 7.0LANWWCDD468 pKa = 3.57FHH470 pKa = 8.0YY471 pKa = 10.75KK472 pKa = 10.53SFISDD477 pKa = 3.2PRR479 pKa = 11.84AKK481 pKa = 10.65VSFNCSFQRR490 pKa = 11.84RR491 pKa = 11.84TGDD494 pKa = 2.73AFTFFGNTLVTMAMFSFCYY513 pKa = 9.21DD514 pKa = 2.85TRR516 pKa = 11.84QFEE519 pKa = 4.46KK520 pKa = 10.52MLFAGDD526 pKa = 3.88DD527 pKa = 3.62SLAISSSPIVGCSDD541 pKa = 3.52YY542 pKa = 10.89FVSLFNMEE550 pKa = 4.67AKK552 pKa = 10.38IMDD555 pKa = 4.49PGVPYY560 pKa = 10.25ICSKK564 pKa = 10.76FLVSDD569 pKa = 3.32EE570 pKa = 4.49LGRR573 pKa = 11.84CFSAPDD579 pKa = 4.72PIRR582 pKa = 11.84EE583 pKa = 3.99FQRR586 pKa = 11.84LGKK589 pKa = 10.39KK590 pKa = 9.87KK591 pKa = 10.21ISADD595 pKa = 3.24NDD597 pKa = 3.52DD598 pKa = 5.32ALFEE602 pKa = 4.29QYY604 pKa = 11.04VGFKK608 pKa = 10.97DD609 pKa = 4.07RR610 pKa = 11.84MSHH613 pKa = 4.76MRR615 pKa = 11.84NFSEE619 pKa = 5.65YY620 pKa = 10.26EE621 pKa = 3.76IQQLKK626 pKa = 10.47IFFNLKK632 pKa = 8.64YY633 pKa = 9.04KK634 pKa = 10.69QSGEE638 pKa = 4.18VIEE641 pKa = 5.83DD642 pKa = 3.47YY643 pKa = 10.7MGACMFYY650 pKa = 10.97SDD652 pKa = 4.0NFKK655 pKa = 10.96NFKK658 pKa = 9.39TLFTKK663 pKa = 9.93TCAPLVAALNKK674 pKa = 9.72RR675 pKa = 11.84VKK677 pKa = 10.4DD678 pKa = 3.54KK679 pKa = 10.85PFRR682 pKa = 11.84LPPSLL687 pKa = 4.07
MM1 pKa = 7.82AAFTFEE7 pKa = 4.28NFLSGAYY14 pKa = 8.81TGLPIDD20 pKa = 3.93KK21 pKa = 10.11FRR23 pKa = 11.84ALGLNTEE30 pKa = 5.06DD31 pKa = 4.17YY32 pKa = 11.27DD33 pKa = 3.88EE34 pKa = 4.7QRR36 pKa = 11.84WEE38 pKa = 4.05MLVKK42 pKa = 10.6CVDD45 pKa = 3.4SGLMQFSCSRR55 pKa = 11.84DD56 pKa = 3.31EE57 pKa = 4.96ALVLLWNEE65 pKa = 3.97EE66 pKa = 4.25EE67 pKa = 4.89LPKK70 pKa = 10.92DD71 pKa = 4.08DD72 pKa = 5.47DD73 pKa = 4.96DD74 pKa = 4.04EE75 pKa = 4.76VPYY78 pKa = 10.13EE79 pKa = 4.42VPCWTPDD86 pKa = 2.94TDD88 pKa = 3.41ATVIDD93 pKa = 5.79DD94 pKa = 3.71VSEE97 pKa = 3.82WLAEE101 pKa = 4.1KK102 pKa = 10.21TSVRR106 pKa = 11.84DD107 pKa = 3.64TVVVCSDD114 pKa = 3.28YY115 pKa = 11.55DD116 pKa = 3.74AVSEE120 pKa = 4.47TPVEE124 pKa = 4.22VLSVEE129 pKa = 4.14LEE131 pKa = 3.97EE132 pKa = 6.24DD133 pKa = 3.78SEE135 pKa = 4.55EE136 pKa = 3.97DD137 pKa = 3.64TIADD141 pKa = 3.34VCLDD145 pKa = 3.28ARR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 9.8SFRR152 pKa = 11.84DD153 pKa = 3.1YY154 pKa = 9.61FTIVEE159 pKa = 4.22EE160 pKa = 4.36EE161 pKa = 4.18FVEE164 pKa = 4.07EE165 pKa = 5.24DD166 pKa = 3.92PLISLNDD173 pKa = 3.66GNVCPVRR180 pKa = 11.84THH182 pKa = 6.17EE183 pKa = 4.33VTSLEE188 pKa = 4.33KK189 pKa = 10.34PVMLDD194 pKa = 2.65VGRR197 pKa = 11.84RR198 pKa = 11.84CDD200 pKa = 3.51RR201 pKa = 11.84VNLEE205 pKa = 4.18SLQGAINMNLPSHH218 pKa = 7.03AYY220 pKa = 10.24FDD222 pKa = 4.69DD223 pKa = 3.38TWHH226 pKa = 6.17QYY228 pKa = 8.96FVEE231 pKa = 4.81GSKK234 pKa = 11.0LDD236 pKa = 3.47VDD238 pKa = 4.86FDD240 pKa = 4.62NIRR243 pKa = 11.84LRR245 pKa = 11.84QSEE248 pKa = 4.72VFCDD252 pKa = 3.68RR253 pKa = 11.84DD254 pKa = 3.14LDD256 pKa = 4.15RR257 pKa = 11.84YY258 pKa = 8.31YY259 pKa = 10.73QPEE262 pKa = 4.46LFAGASSRR270 pKa = 11.84RR271 pKa = 11.84IGTQKK276 pKa = 10.21EE277 pKa = 3.7ALVAIRR283 pKa = 11.84KK284 pKa = 9.45RR285 pKa = 11.84NADD288 pKa = 3.54VPEE291 pKa = 4.47LADD294 pKa = 3.59SVDD297 pKa = 3.52VEE299 pKa = 4.29RR300 pKa = 11.84LSEE303 pKa = 4.23SVAKK307 pKa = 10.37KK308 pKa = 9.94FLSSYY313 pKa = 10.69VCDD316 pKa = 4.51LKK318 pKa = 11.16PVVGVMEE325 pKa = 4.54KK326 pKa = 9.75MRR328 pKa = 11.84AYY330 pKa = 8.88HH331 pKa = 6.01QKK333 pKa = 9.6WGDD336 pKa = 3.54KK337 pKa = 9.49IDD339 pKa = 3.6PMFLLKK345 pKa = 10.27EE346 pKa = 4.29HH347 pKa = 6.39NLQRR351 pKa = 11.84YY352 pKa = 5.82EE353 pKa = 4.23HH354 pKa = 6.57MIKK357 pKa = 9.59TDD359 pKa = 3.37VKK361 pKa = 8.52PTVAHH366 pKa = 5.81SMHH369 pKa = 6.59VEE371 pKa = 3.75RR372 pKa = 11.84AIPATITFHH381 pKa = 6.56GKK383 pKa = 9.69SICAGFSPSFTALFDD398 pKa = 3.86EE399 pKa = 5.18FQKK402 pKa = 11.26SLDD405 pKa = 3.69EE406 pKa = 4.08RR407 pKa = 11.84VVIPSGPISTIEE419 pKa = 3.64MDD421 pKa = 3.65FDD423 pKa = 3.66IRR425 pKa = 11.84NKK427 pKa = 10.67YY428 pKa = 8.51YY429 pKa = 11.19LEE431 pKa = 4.25VDD433 pKa = 3.8LSKK436 pKa = 10.71FDD438 pKa = 3.62KK439 pKa = 11.14SQGLLHH445 pKa = 7.4LEE447 pKa = 4.19FQRR450 pKa = 11.84KK451 pKa = 7.97ILCKK455 pKa = 9.97IGLPAHH461 pKa = 7.0LANWWCDD468 pKa = 3.57FHH470 pKa = 8.0YY471 pKa = 10.75KK472 pKa = 10.53SFISDD477 pKa = 3.2PRR479 pKa = 11.84AKK481 pKa = 10.65VSFNCSFQRR490 pKa = 11.84RR491 pKa = 11.84TGDD494 pKa = 2.73AFTFFGNTLVTMAMFSFCYY513 pKa = 9.21DD514 pKa = 2.85TRR516 pKa = 11.84QFEE519 pKa = 4.46KK520 pKa = 10.52MLFAGDD526 pKa = 3.88DD527 pKa = 3.62SLAISSSPIVGCSDD541 pKa = 3.52YY542 pKa = 10.89FVSLFNMEE550 pKa = 4.67AKK552 pKa = 10.38IMDD555 pKa = 4.49PGVPYY560 pKa = 10.25ICSKK564 pKa = 10.76FLVSDD569 pKa = 3.32EE570 pKa = 4.49LGRR573 pKa = 11.84CFSAPDD579 pKa = 4.72PIRR582 pKa = 11.84EE583 pKa = 3.99FQRR586 pKa = 11.84LGKK589 pKa = 10.39KK590 pKa = 9.87KK591 pKa = 10.21ISADD595 pKa = 3.24NDD597 pKa = 3.52DD598 pKa = 5.32ALFEE602 pKa = 4.29QYY604 pKa = 11.04VGFKK608 pKa = 10.97DD609 pKa = 4.07RR610 pKa = 11.84MSHH613 pKa = 4.76MRR615 pKa = 11.84NFSEE619 pKa = 5.65YY620 pKa = 10.26EE621 pKa = 3.76IQQLKK626 pKa = 10.47IFFNLKK632 pKa = 8.64YY633 pKa = 9.04KK634 pKa = 10.69QSGEE638 pKa = 4.18VIEE641 pKa = 5.83DD642 pKa = 3.47YY643 pKa = 10.7MGACMFYY650 pKa = 10.97SDD652 pKa = 4.0NFKK655 pKa = 10.96NFKK658 pKa = 9.39TLFTKK663 pKa = 9.93TCAPLVAALNKK674 pKa = 9.72RR675 pKa = 11.84VKK677 pKa = 10.4DD678 pKa = 3.54KK679 pKa = 10.85PFRR682 pKa = 11.84LPPSLL687 pKa = 4.07
Molecular weight: 79.0 kDa
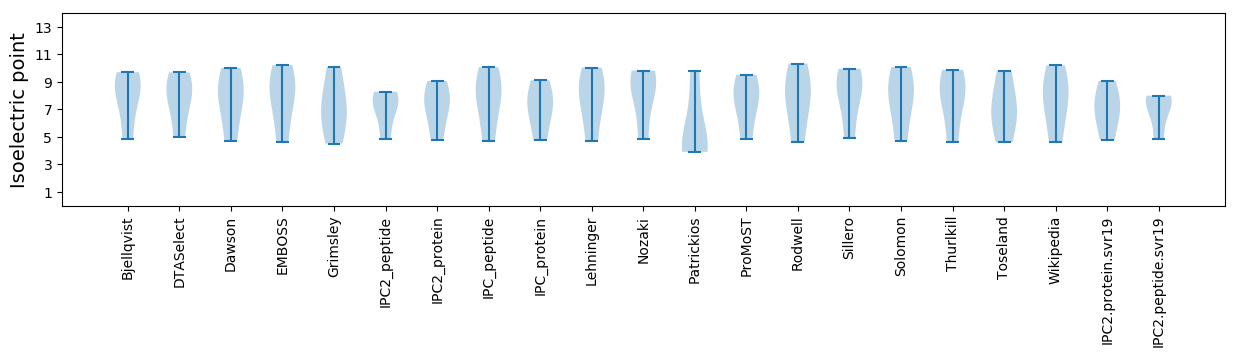
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9DUT1|MVP_PZSVT Movement protein OS=Pelargonium zonate spot virus (isolate Tomato/Italy/1982) OX=650488 GN=ORF3a PE=3 SV=1
MM1 pKa = 7.5PPKK4 pKa = 10.21RR5 pKa = 11.84QNTEE9 pKa = 3.34TRR11 pKa = 11.84KK12 pKa = 9.75ARR14 pKa = 11.84QNRR17 pKa = 11.84ARR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84QQALAKK28 pKa = 9.98LARR31 pKa = 11.84EE32 pKa = 4.16FSGLSMSVEE41 pKa = 4.35RR42 pKa = 11.84SPSTSWADD50 pKa = 3.09ITEE53 pKa = 4.32SEE55 pKa = 4.55SRR57 pKa = 11.84LKK59 pKa = 10.78LIPGFTATEE68 pKa = 4.28VTFDD72 pKa = 3.61PSLTFGTHH80 pKa = 4.88TGFATAEE87 pKa = 4.02RR88 pKa = 11.84SLTVPDD94 pKa = 4.96ALLEE98 pKa = 4.41SPNLRR103 pKa = 11.84LNRR106 pKa = 11.84VAVVVLLDD114 pKa = 3.55PTVPEE119 pKa = 4.13AHH121 pKa = 7.02KK122 pKa = 9.92FWCALGDD129 pKa = 3.35RR130 pKa = 11.84WVAPSVGSFPSNAVRR145 pKa = 11.84ITGRR149 pKa = 11.84EE150 pKa = 4.01GKK152 pKa = 10.22GHH154 pKa = 6.9VIYY157 pKa = 9.91HH158 pKa = 5.32YY159 pKa = 9.94PGKK162 pKa = 8.57TVEE165 pKa = 4.57HH166 pKa = 6.32LAKK169 pKa = 10.6LRR171 pKa = 11.84VYY173 pKa = 10.77LFATDD178 pKa = 3.67YY179 pKa = 11.46AIVGNNSPVATVKK192 pKa = 10.54IFVEE196 pKa = 4.34HH197 pKa = 6.35EE198 pKa = 4.22KK199 pKa = 10.33IGQAEE204 pKa = 4.55YY205 pKa = 10.66IPLL208 pKa = 4.2
MM1 pKa = 7.5PPKK4 pKa = 10.21RR5 pKa = 11.84QNTEE9 pKa = 3.34TRR11 pKa = 11.84KK12 pKa = 9.75ARR14 pKa = 11.84QNRR17 pKa = 11.84ARR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84QQALAKK28 pKa = 9.98LARR31 pKa = 11.84EE32 pKa = 4.16FSGLSMSVEE41 pKa = 4.35RR42 pKa = 11.84SPSTSWADD50 pKa = 3.09ITEE53 pKa = 4.32SEE55 pKa = 4.55SRR57 pKa = 11.84LKK59 pKa = 10.78LIPGFTATEE68 pKa = 4.28VTFDD72 pKa = 3.61PSLTFGTHH80 pKa = 4.88TGFATAEE87 pKa = 4.02RR88 pKa = 11.84SLTVPDD94 pKa = 4.96ALLEE98 pKa = 4.41SPNLRR103 pKa = 11.84LNRR106 pKa = 11.84VAVVVLLDD114 pKa = 3.55PTVPEE119 pKa = 4.13AHH121 pKa = 7.02KK122 pKa = 9.92FWCALGDD129 pKa = 3.35RR130 pKa = 11.84WVAPSVGSFPSNAVRR145 pKa = 11.84ITGRR149 pKa = 11.84EE150 pKa = 4.01GKK152 pKa = 10.22GHH154 pKa = 6.9VIYY157 pKa = 9.91HH158 pKa = 5.32YY159 pKa = 9.94PGKK162 pKa = 8.57TVEE165 pKa = 4.57HH166 pKa = 6.32LAKK169 pKa = 10.6LRR171 pKa = 11.84VYY173 pKa = 10.77LFATDD178 pKa = 3.67YY179 pKa = 11.46AIVGNNSPVATVKK192 pKa = 10.54IFVEE196 pKa = 4.34HH197 pKa = 6.35EE198 pKa = 4.22KK199 pKa = 10.33IGQAEE204 pKa = 4.55YY205 pKa = 10.66IPLL208 pKa = 4.2
Molecular weight: 23.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2166 |
208 |
962 |
541.5 |
61.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.233 ± 0.569 | 2.493 ± 0.345 |
6.464 ± 1.149 | 6.648 ± 0.383 |
5.448 ± 0.696 | 5.91 ± 0.833 |
2.401 ± 0.254 | 4.294 ± 0.203 |
6.371 ± 0.377 | 8.68 ± 0.287 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.139 ± 0.43 | 3.555 ± 0.174 |
4.294 ± 0.624 | 2.77 ± 0.165 |
5.91 ± 0.372 | 8.495 ± 0.49 |
5.171 ± 0.535 | 7.756 ± 0.146 |
1.293 ± 0.1 | 2.678 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
