
Catenaria anguillulae PL171
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Blastocladiomycota; Blastocladiomycota incertae sedis; Blastocladiomycetes; Blastocladiales; Catenariaceae; Catenaria; Catenaria anguillulae
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
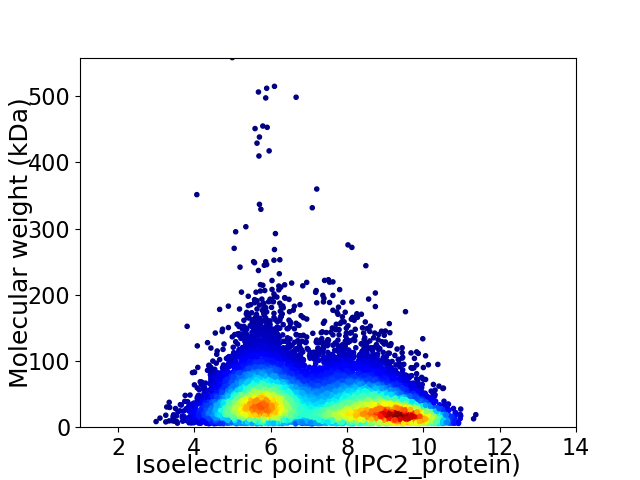
Virtual 2D-PAGE plot for 12599 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y2HUA0|A0A1Y2HUA0_9FUNG Uncharacterized protein OS=Catenaria anguillulae PL171 OX=765915 GN=BCR44DRAFT_41493 PE=4 SV=1
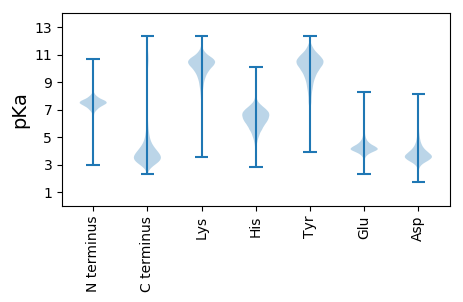
MM1 pKa = 7.15SAAAAYY7 pKa = 9.95YY8 pKa = 8.25PLPCHH13 pKa = 6.04STDD16 pKa = 3.26ITAQTQAIDD25 pKa = 3.45QSQVANAGDD34 pKa = 3.63NSVIIQIQQAVQAQKK49 pKa = 11.47ADD51 pKa = 3.74VASTAAAGGRR61 pKa = 11.84HH62 pKa = 5.42MSDD65 pKa = 3.51TYY67 pKa = 11.25DD68 pKa = 3.36QQDD71 pKa = 3.42SVAQSQDD78 pKa = 3.08LNQVQFANSGDD89 pKa = 3.6NSVVIQNQNAVQSQFGNLDD108 pKa = 3.27SSDD111 pKa = 3.23GHH113 pKa = 6.38GWATGNSIDD122 pKa = 4.92QAQDD126 pKa = 3.12LNQFQFANASDD137 pKa = 3.55NSINQNAVQNQQANVGGGRR156 pKa = 11.84HH157 pKa = 4.88MHH159 pKa = 7.17DD160 pKa = 4.12ANTVAQAQDD169 pKa = 3.55LNQLQFADD177 pKa = 4.31AGDD180 pKa = 3.66NSAIIQNQDD189 pKa = 2.69APQSQFANIHH199 pKa = 6.3DD200 pKa = 4.37FGGHH204 pKa = 6.39GYY206 pKa = 10.49GHH208 pKa = 6.93TGYY211 pKa = 8.96PDD213 pKa = 3.69SMITQAQDD221 pKa = 3.08LTQLQFANAGDD232 pKa = 3.72NAIIVQNQDD241 pKa = 2.57AFQSQFANLFGGRR254 pKa = 11.84HH255 pKa = 5.0TSDD258 pKa = 3.8ADD260 pKa = 4.38VIAQAQNLDD269 pKa = 3.27QFQFANAGDD278 pKa = 4.03HH279 pKa = 6.85SAVLQNQDD287 pKa = 3.13ALQSQFALTNVDD299 pKa = 3.14PHH301 pKa = 7.8GYY303 pKa = 9.25AASGGAGHH311 pKa = 7.57AGGSDD316 pKa = 2.84WLATFDD322 pKa = 4.42FTHH325 pKa = 6.04QVQDD329 pKa = 3.76LNQVQFASAAGGDD342 pKa = 3.8GGASAMHH349 pKa = 6.87ASVMPDD355 pKa = 2.86VTEE358 pKa = 4.21PLVTMSLFF366 pKa = 3.55
MM1 pKa = 7.15SAAAAYY7 pKa = 9.95YY8 pKa = 8.25PLPCHH13 pKa = 6.04STDD16 pKa = 3.26ITAQTQAIDD25 pKa = 3.45QSQVANAGDD34 pKa = 3.63NSVIIQIQQAVQAQKK49 pKa = 11.47ADD51 pKa = 3.74VASTAAAGGRR61 pKa = 11.84HH62 pKa = 5.42MSDD65 pKa = 3.51TYY67 pKa = 11.25DD68 pKa = 3.36QQDD71 pKa = 3.42SVAQSQDD78 pKa = 3.08LNQVQFANSGDD89 pKa = 3.6NSVVIQNQNAVQSQFGNLDD108 pKa = 3.27SSDD111 pKa = 3.23GHH113 pKa = 6.38GWATGNSIDD122 pKa = 4.92QAQDD126 pKa = 3.12LNQFQFANASDD137 pKa = 3.55NSINQNAVQNQQANVGGGRR156 pKa = 11.84HH157 pKa = 4.88MHH159 pKa = 7.17DD160 pKa = 4.12ANTVAQAQDD169 pKa = 3.55LNQLQFADD177 pKa = 4.31AGDD180 pKa = 3.66NSAIIQNQDD189 pKa = 2.69APQSQFANIHH199 pKa = 6.3DD200 pKa = 4.37FGGHH204 pKa = 6.39GYY206 pKa = 10.49GHH208 pKa = 6.93TGYY211 pKa = 8.96PDD213 pKa = 3.69SMITQAQDD221 pKa = 3.08LTQLQFANAGDD232 pKa = 3.72NAIIVQNQDD241 pKa = 2.57AFQSQFANLFGGRR254 pKa = 11.84HH255 pKa = 5.0TSDD258 pKa = 3.8ADD260 pKa = 4.38VIAQAQNLDD269 pKa = 3.27QFQFANAGDD278 pKa = 4.03HH279 pKa = 6.85SAVLQNQDD287 pKa = 3.13ALQSQFALTNVDD299 pKa = 3.14PHH301 pKa = 7.8GYY303 pKa = 9.25AASGGAGHH311 pKa = 7.57AGGSDD316 pKa = 2.84WLATFDD322 pKa = 4.42FTHH325 pKa = 6.04QVQDD329 pKa = 3.76LNQVQFASAAGGDD342 pKa = 3.8GGASAMHH349 pKa = 6.87ASVMPDD355 pKa = 2.86VTEE358 pKa = 4.21PLVTMSLFF366 pKa = 3.55
Molecular weight: 38.53 kDa
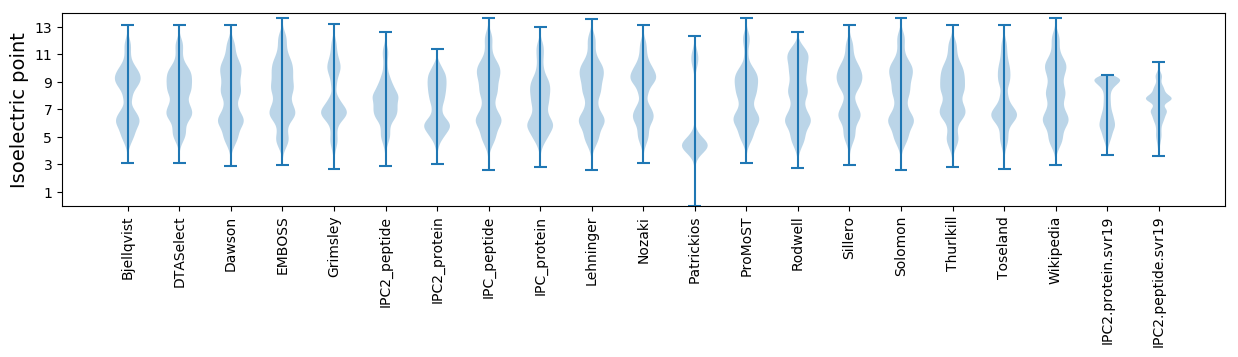
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y2I6M2|A0A1Y2I6M2_9FUNG RRM domain-containing protein OS=Catenaria anguillulae PL171 OX=765915 GN=BCR44DRAFT_122977 PE=4 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.74RR7 pKa = 11.84PRR9 pKa = 11.84QQSRR13 pKa = 11.84AMRR16 pKa = 11.84RR17 pKa = 11.84QPKK20 pKa = 10.01LLRR23 pKa = 11.84QPSRR27 pKa = 11.84ATRR30 pKa = 11.84NPPRR34 pKa = 11.84LRR36 pKa = 11.84RR37 pKa = 11.84QPNLVMRR44 pKa = 11.84NQRR47 pKa = 11.84RR48 pKa = 11.84LLRR51 pKa = 11.84QPSRR55 pKa = 11.84AMRR58 pKa = 11.84NQPRR62 pKa = 11.84RR63 pKa = 11.84LHH65 pKa = 5.82QPSQEE70 pKa = 4.25TTSNQPRR77 pKa = 11.84LPHH80 pKa = 5.41QPSRR84 pKa = 11.84VTMQHH89 pKa = 6.06PSKK92 pKa = 10.51RR93 pKa = 11.84PPQPKK98 pKa = 8.78PAKK101 pKa = 6.61TTSPP105 pKa = 3.49
MM1 pKa = 7.48RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.74RR7 pKa = 11.84PRR9 pKa = 11.84QQSRR13 pKa = 11.84AMRR16 pKa = 11.84RR17 pKa = 11.84QPKK20 pKa = 10.01LLRR23 pKa = 11.84QPSRR27 pKa = 11.84ATRR30 pKa = 11.84NPPRR34 pKa = 11.84LRR36 pKa = 11.84RR37 pKa = 11.84QPNLVMRR44 pKa = 11.84NQRR47 pKa = 11.84RR48 pKa = 11.84LLRR51 pKa = 11.84QPSRR55 pKa = 11.84AMRR58 pKa = 11.84NQPRR62 pKa = 11.84RR63 pKa = 11.84LHH65 pKa = 5.82QPSQEE70 pKa = 4.25TTSNQPRR77 pKa = 11.84LPHH80 pKa = 5.41QPSRR84 pKa = 11.84VTMQHH89 pKa = 6.06PSKK92 pKa = 10.51RR93 pKa = 11.84PPQPKK98 pKa = 8.78PAKK101 pKa = 6.61TTSPP105 pKa = 3.49
Molecular weight: 12.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4929253 |
49 |
5022 |
391.2 |
42.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.969 ± 0.029 | 1.443 ± 0.011 |
5.267 ± 0.018 | 4.509 ± 0.023 |
3.252 ± 0.015 | 6.533 ± 0.021 |
2.959 ± 0.013 | 3.908 ± 0.016 |
3.869 ± 0.022 | 9.305 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.376 ± 0.01 | 3.033 ± 0.012 |
6.816 ± 0.03 | 4.079 ± 0.017 |
6.211 ± 0.023 | 8.717 ± 0.032 |
6.163 ± 0.016 | 7.087 ± 0.019 |
1.352 ± 0.009 | 2.154 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
