
Lachnotalea glycerini
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnotalea
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
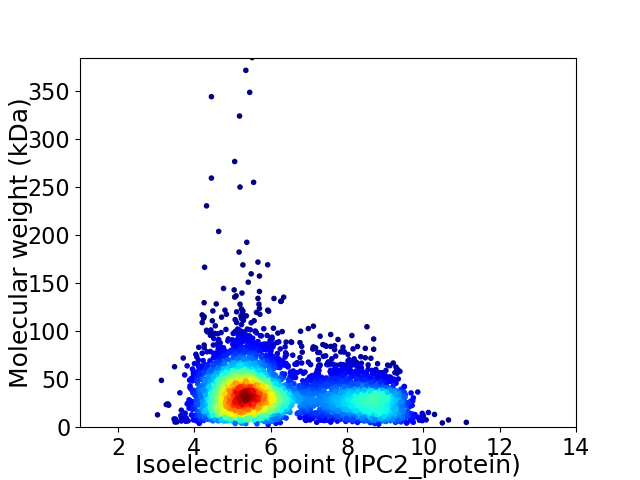
Virtual 2D-PAGE plot for 4293 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A371JI86|A0A371JI86_9FIRM Uncharacterized protein OS=Lachnotalea glycerini OX=1763509 GN=CG710_005060 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.23FKK4 pKa = 11.19NFLAVSLAFTMVAGTLSGCSSEE26 pKa = 4.4TGGEE30 pKa = 4.15VASAAVTNQGKK41 pKa = 7.35TMEE44 pKa = 4.38YY45 pKa = 9.92EE46 pKa = 4.17SKK48 pKa = 10.97LFGDD52 pKa = 5.06DD53 pKa = 4.61PITVNIIVDD62 pKa = 3.58EE63 pKa = 4.58TEE65 pKa = 3.88WQNMIDD71 pKa = 3.64NMSDD75 pKa = 3.1EE76 pKa = 4.95EE77 pKa = 4.52YY78 pKa = 10.37ISVNMEE84 pKa = 3.76VNNEE88 pKa = 4.06LFSDD92 pKa = 3.79VAIRR96 pKa = 11.84AKK98 pKa = 10.7GNSSLRR104 pKa = 11.84SVSEE108 pKa = 4.04SDD110 pKa = 2.88SDD112 pKa = 3.69RR113 pKa = 11.84YY114 pKa = 10.05SFKK117 pKa = 11.37VKK119 pKa = 10.16FDD121 pKa = 3.99KK122 pKa = 10.81YY123 pKa = 10.88QKK125 pKa = 10.45NQTCYY130 pKa = 11.05GLDD133 pKa = 3.25KK134 pKa = 11.09LVLNNVISDD143 pKa = 3.7NTYY146 pKa = 8.64MKK148 pKa = 11.2DD149 pKa = 3.26MIAFDD154 pKa = 3.41MMNYY158 pKa = 9.36IGAYY162 pKa = 9.08SSLYY166 pKa = 8.45NYY168 pKa = 10.89AVISVNGEE176 pKa = 3.56YY177 pKa = 9.6WGLYY181 pKa = 8.46IALEE185 pKa = 4.91GYY187 pKa = 10.37DD188 pKa = 4.05SSFCDD193 pKa = 3.32RR194 pKa = 11.84VFGNDD199 pKa = 4.21DD200 pKa = 3.48GNLYY204 pKa = 8.79NVKK207 pKa = 9.29MVNNGGGGPGGPDD220 pKa = 3.11QSNVDD225 pKa = 3.37GQAADD230 pKa = 3.39VSGTADD236 pKa = 3.3TQTTEE241 pKa = 4.06LTTGSDD247 pKa = 3.38TQTTDD252 pKa = 2.49SAQAQDD258 pKa = 3.7TTNEE262 pKa = 4.04NQAEE266 pKa = 4.17PPEE269 pKa = 4.18MQGGNEE275 pKa = 4.17PPSDD279 pKa = 3.84LTSALPDD286 pKa = 3.68GFDD289 pKa = 3.12IDD291 pKa = 5.73DD292 pKa = 5.24YY293 pKa = 9.6ITKK296 pKa = 10.23SAEE299 pKa = 3.89EE300 pKa = 4.4GFSLSSYY307 pKa = 10.79LEE309 pKa = 3.96EE310 pKa = 6.13LGLTEE315 pKa = 5.53ADD317 pKa = 4.27FSIPSNMDD325 pKa = 3.31LEE327 pKa = 5.78DD328 pKa = 3.77MLDD331 pKa = 3.27MVVAIKK337 pKa = 10.93NGDD340 pKa = 3.5FSDD343 pKa = 3.63EE344 pKa = 4.32SMGGGKK350 pKa = 10.38GGGGMGGSSTGGALLYY366 pKa = 10.35TDD368 pKa = 5.8DD369 pKa = 5.36EE370 pKa = 4.58ISSYY374 pKa = 11.62SSIFEE379 pKa = 3.89NTSFNHH385 pKa = 4.87TTEE388 pKa = 5.29DD389 pKa = 3.38DD390 pKa = 3.44QKK392 pKa = 11.43RR393 pKa = 11.84VIEE396 pKa = 4.79AIKK399 pKa = 10.54ALNAGEE405 pKa = 5.19NIDD408 pKa = 3.52TYY410 pKa = 11.49FNVDD414 pKa = 3.43EE415 pKa = 4.31ILRR418 pKa = 11.84YY419 pKa = 9.22LAAHH423 pKa = 5.97TVLCNFDD430 pKa = 4.3SYY432 pKa = 11.34SSSMAQNYY440 pKa = 9.04YY441 pKa = 9.99IYY443 pKa = 10.65EE444 pKa = 4.33EE445 pKa = 4.4DD446 pKa = 3.6SKK448 pKa = 11.48ISILPWDD455 pKa = 3.77YY456 pKa = 11.57NLAFGGFQSEE466 pKa = 4.73SASQCVNWAIDD477 pKa = 3.83TPVLGVDD484 pKa = 3.22MADD487 pKa = 3.51RR488 pKa = 11.84PLINQLLSNEE498 pKa = 4.49EE499 pKa = 3.99YY500 pKa = 10.3LATYY504 pKa = 10.14HH505 pKa = 7.33EE506 pKa = 4.69YY507 pKa = 10.66LQQLVDD513 pKa = 4.01GYY515 pKa = 9.14MANFSEE521 pKa = 5.04HH522 pKa = 5.87VDD524 pKa = 3.58NLVSKK529 pKa = 10.32IDD531 pKa = 4.07SYY533 pKa = 11.95VQTDD537 pKa = 3.49PTAFCTYY544 pKa = 10.31EE545 pKa = 4.01EE546 pKa = 4.5YY547 pKa = 9.31QTAVSTLKK555 pKa = 10.91SFMEE559 pKa = 4.37LRR561 pKa = 11.84MQSIQGQLDD570 pKa = 3.44GTIPSTTQGQEE581 pKa = 3.71QDD583 pKa = 3.05SSTLIDD589 pKa = 3.83ASAITLSDD597 pKa = 3.3MGSQGGGKK605 pKa = 8.04DD606 pKa = 3.35QKK608 pKa = 11.39GDD610 pKa = 3.39MPGRR614 pKa = 11.84PGQNDD619 pKa = 3.57ANTSTEE625 pKa = 4.06TNADD629 pKa = 3.33GTGDD633 pKa = 3.76TEE635 pKa = 4.57TTSDD639 pKa = 3.83TEE641 pKa = 4.52TTSDD645 pKa = 3.71TEE647 pKa = 4.55TTSDD651 pKa = 3.27TGTTSDD657 pKa = 3.67TGTTSAQQ664 pKa = 3.13
MM1 pKa = 7.48KK2 pKa = 10.23FKK4 pKa = 11.19NFLAVSLAFTMVAGTLSGCSSEE26 pKa = 4.4TGGEE30 pKa = 4.15VASAAVTNQGKK41 pKa = 7.35TMEE44 pKa = 4.38YY45 pKa = 9.92EE46 pKa = 4.17SKK48 pKa = 10.97LFGDD52 pKa = 5.06DD53 pKa = 4.61PITVNIIVDD62 pKa = 3.58EE63 pKa = 4.58TEE65 pKa = 3.88WQNMIDD71 pKa = 3.64NMSDD75 pKa = 3.1EE76 pKa = 4.95EE77 pKa = 4.52YY78 pKa = 10.37ISVNMEE84 pKa = 3.76VNNEE88 pKa = 4.06LFSDD92 pKa = 3.79VAIRR96 pKa = 11.84AKK98 pKa = 10.7GNSSLRR104 pKa = 11.84SVSEE108 pKa = 4.04SDD110 pKa = 2.88SDD112 pKa = 3.69RR113 pKa = 11.84YY114 pKa = 10.05SFKK117 pKa = 11.37VKK119 pKa = 10.16FDD121 pKa = 3.99KK122 pKa = 10.81YY123 pKa = 10.88QKK125 pKa = 10.45NQTCYY130 pKa = 11.05GLDD133 pKa = 3.25KK134 pKa = 11.09LVLNNVISDD143 pKa = 3.7NTYY146 pKa = 8.64MKK148 pKa = 11.2DD149 pKa = 3.26MIAFDD154 pKa = 3.41MMNYY158 pKa = 9.36IGAYY162 pKa = 9.08SSLYY166 pKa = 8.45NYY168 pKa = 10.89AVISVNGEE176 pKa = 3.56YY177 pKa = 9.6WGLYY181 pKa = 8.46IALEE185 pKa = 4.91GYY187 pKa = 10.37DD188 pKa = 4.05SSFCDD193 pKa = 3.32RR194 pKa = 11.84VFGNDD199 pKa = 4.21DD200 pKa = 3.48GNLYY204 pKa = 8.79NVKK207 pKa = 9.29MVNNGGGGPGGPDD220 pKa = 3.11QSNVDD225 pKa = 3.37GQAADD230 pKa = 3.39VSGTADD236 pKa = 3.3TQTTEE241 pKa = 4.06LTTGSDD247 pKa = 3.38TQTTDD252 pKa = 2.49SAQAQDD258 pKa = 3.7TTNEE262 pKa = 4.04NQAEE266 pKa = 4.17PPEE269 pKa = 4.18MQGGNEE275 pKa = 4.17PPSDD279 pKa = 3.84LTSALPDD286 pKa = 3.68GFDD289 pKa = 3.12IDD291 pKa = 5.73DD292 pKa = 5.24YY293 pKa = 9.6ITKK296 pKa = 10.23SAEE299 pKa = 3.89EE300 pKa = 4.4GFSLSSYY307 pKa = 10.79LEE309 pKa = 3.96EE310 pKa = 6.13LGLTEE315 pKa = 5.53ADD317 pKa = 4.27FSIPSNMDD325 pKa = 3.31LEE327 pKa = 5.78DD328 pKa = 3.77MLDD331 pKa = 3.27MVVAIKK337 pKa = 10.93NGDD340 pKa = 3.5FSDD343 pKa = 3.63EE344 pKa = 4.32SMGGGKK350 pKa = 10.38GGGGMGGSSTGGALLYY366 pKa = 10.35TDD368 pKa = 5.8DD369 pKa = 5.36EE370 pKa = 4.58ISSYY374 pKa = 11.62SSIFEE379 pKa = 3.89NTSFNHH385 pKa = 4.87TTEE388 pKa = 5.29DD389 pKa = 3.38DD390 pKa = 3.44QKK392 pKa = 11.43RR393 pKa = 11.84VIEE396 pKa = 4.79AIKK399 pKa = 10.54ALNAGEE405 pKa = 5.19NIDD408 pKa = 3.52TYY410 pKa = 11.49FNVDD414 pKa = 3.43EE415 pKa = 4.31ILRR418 pKa = 11.84YY419 pKa = 9.22LAAHH423 pKa = 5.97TVLCNFDD430 pKa = 4.3SYY432 pKa = 11.34SSSMAQNYY440 pKa = 9.04YY441 pKa = 9.99IYY443 pKa = 10.65EE444 pKa = 4.33EE445 pKa = 4.4DD446 pKa = 3.6SKK448 pKa = 11.48ISILPWDD455 pKa = 3.77YY456 pKa = 11.57NLAFGGFQSEE466 pKa = 4.73SASQCVNWAIDD477 pKa = 3.83TPVLGVDD484 pKa = 3.22MADD487 pKa = 3.51RR488 pKa = 11.84PLINQLLSNEE498 pKa = 4.49EE499 pKa = 3.99YY500 pKa = 10.3LATYY504 pKa = 10.14HH505 pKa = 7.33EE506 pKa = 4.69YY507 pKa = 10.66LQQLVDD513 pKa = 4.01GYY515 pKa = 9.14MANFSEE521 pKa = 5.04HH522 pKa = 5.87VDD524 pKa = 3.58NLVSKK529 pKa = 10.32IDD531 pKa = 4.07SYY533 pKa = 11.95VQTDD537 pKa = 3.49PTAFCTYY544 pKa = 10.31EE545 pKa = 4.01EE546 pKa = 4.5YY547 pKa = 9.31QTAVSTLKK555 pKa = 10.91SFMEE559 pKa = 4.37LRR561 pKa = 11.84MQSIQGQLDD570 pKa = 3.44GTIPSTTQGQEE581 pKa = 3.71QDD583 pKa = 3.05SSTLIDD589 pKa = 3.83ASAITLSDD597 pKa = 3.3MGSQGGGKK605 pKa = 8.04DD606 pKa = 3.35QKK608 pKa = 11.39GDD610 pKa = 3.39MPGRR614 pKa = 11.84PGQNDD619 pKa = 3.57ANTSTEE625 pKa = 4.06TNADD629 pKa = 3.33GTGDD633 pKa = 3.76TEE635 pKa = 4.57TTSDD639 pKa = 3.83TEE641 pKa = 4.52TTSDD645 pKa = 3.71TEE647 pKa = 4.55TTSDD651 pKa = 3.27TGTTSDD657 pKa = 3.67TGTTSAQQ664 pKa = 3.13
Molecular weight: 71.99 kDa
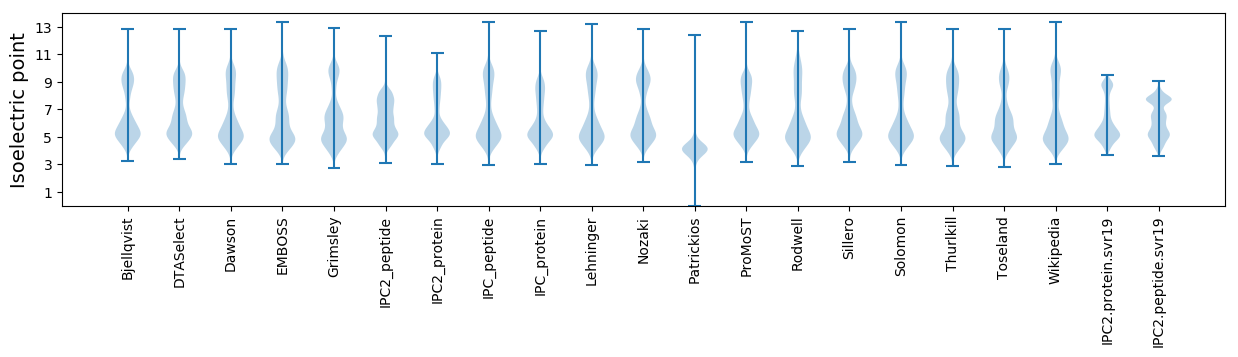
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A255I8D4|A0A255I8D4_9FIRM Zinc metalloprotease OS=Lachnotalea glycerini OX=1763509 GN=rseP PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1421011 |
25 |
3375 |
331.0 |
37.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.425 ± 0.039 | 1.392 ± 0.014 |
5.386 ± 0.029 | 7.257 ± 0.046 |
4.272 ± 0.029 | 6.322 ± 0.033 |
1.566 ± 0.016 | 9.124 ± 0.045 |
7.675 ± 0.032 | 8.979 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.944 ± 0.019 | 5.624 ± 0.037 |
2.76 ± 0.02 | 3.276 ± 0.019 |
3.369 ± 0.025 | 6.415 ± 0.034 |
5.502 ± 0.041 | 6.38 ± 0.026 |
0.86 ± 0.013 | 4.473 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
