
Elm carlavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus; unclassified Carlavirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
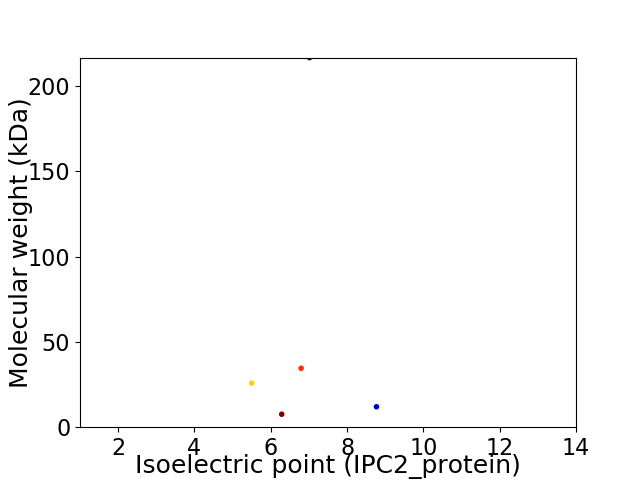
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R7Z6N6|A0A2R7Z6N6_9VIRU 7 kDa protein OS=Elm carlavirus OX=2015551 GN=ORF4 PE=3 SV=1
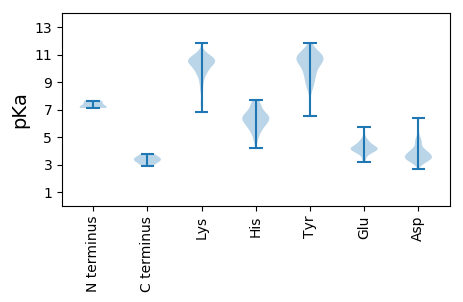
MM1 pKa = 7.54NSVLDD6 pKa = 3.77VLCEE10 pKa = 4.13FGFSRR15 pKa = 11.84ISISSEE21 pKa = 3.58RR22 pKa = 11.84PLVVHH27 pKa = 6.5CVPGAGKK34 pKa = 8.41STAIRR39 pKa = 11.84SILRR43 pKa = 11.84SCEE46 pKa = 3.47NSVAYY51 pKa = 9.19TFGVCDD57 pKa = 3.4PAQIEE62 pKa = 4.68GCRR65 pKa = 11.84IRR67 pKa = 11.84SASEE71 pKa = 3.17WHH73 pKa = 6.63EE74 pKa = 4.43PNSGTIVIVDD84 pKa = 4.73EE85 pKa = 4.2YY86 pKa = 11.22TEE88 pKa = 4.77GDD90 pKa = 3.98FKK92 pKa = 11.22SLKK95 pKa = 7.52PTVVFGDD102 pKa = 3.99PCQAPVSAITLKK114 pKa = 10.61PHH116 pKa = 7.19FICKK120 pKa = 6.83STKK123 pKa = 9.57RR124 pKa = 11.84FGKK127 pKa = 7.35EE128 pKa = 3.33TCNLLRR134 pKa = 11.84SLGYY138 pKa = 10.11EE139 pKa = 3.81INSEE143 pKa = 4.01KK144 pKa = 10.9GDD146 pKa = 3.96TLVHH150 pKa = 5.81EE151 pKa = 5.33PIFEE155 pKa = 4.42GEE157 pKa = 4.09LEE159 pKa = 4.18GVIVAFEE166 pKa = 4.2SDD168 pKa = 4.0VKK170 pKa = 11.03HH171 pKa = 6.34LLAEE175 pKa = 4.89HH176 pKa = 6.07YY177 pKa = 10.85CEE179 pKa = 5.41FLDD182 pKa = 3.6ICEE185 pKa = 4.15IRR187 pKa = 11.84GKK189 pKa = 8.06TFEE192 pKa = 4.16VVTFVTSTEE201 pKa = 4.08GKK203 pKa = 8.91PVKK206 pKa = 10.36NLEE209 pKa = 4.37KK210 pKa = 10.58YY211 pKa = 7.39YY212 pKa = 10.71QCLTRR217 pKa = 11.84HH218 pKa = 5.03SSKK221 pKa = 10.99LKK223 pKa = 10.28ILSPDD228 pKa = 2.96ASFASSS234 pKa = 2.9
MM1 pKa = 7.54NSVLDD6 pKa = 3.77VLCEE10 pKa = 4.13FGFSRR15 pKa = 11.84ISISSEE21 pKa = 3.58RR22 pKa = 11.84PLVVHH27 pKa = 6.5CVPGAGKK34 pKa = 8.41STAIRR39 pKa = 11.84SILRR43 pKa = 11.84SCEE46 pKa = 3.47NSVAYY51 pKa = 9.19TFGVCDD57 pKa = 3.4PAQIEE62 pKa = 4.68GCRR65 pKa = 11.84IRR67 pKa = 11.84SASEE71 pKa = 3.17WHH73 pKa = 6.63EE74 pKa = 4.43PNSGTIVIVDD84 pKa = 4.73EE85 pKa = 4.2YY86 pKa = 11.22TEE88 pKa = 4.77GDD90 pKa = 3.98FKK92 pKa = 11.22SLKK95 pKa = 7.52PTVVFGDD102 pKa = 3.99PCQAPVSAITLKK114 pKa = 10.61PHH116 pKa = 7.19FICKK120 pKa = 6.83STKK123 pKa = 9.57RR124 pKa = 11.84FGKK127 pKa = 7.35EE128 pKa = 3.33TCNLLRR134 pKa = 11.84SLGYY138 pKa = 10.11EE139 pKa = 3.81INSEE143 pKa = 4.01KK144 pKa = 10.9GDD146 pKa = 3.96TLVHH150 pKa = 5.81EE151 pKa = 5.33PIFEE155 pKa = 4.42GEE157 pKa = 4.09LEE159 pKa = 4.18GVIVAFEE166 pKa = 4.2SDD168 pKa = 4.0VKK170 pKa = 11.03HH171 pKa = 6.34LLAEE175 pKa = 4.89HH176 pKa = 6.07YY177 pKa = 10.85CEE179 pKa = 5.41FLDD182 pKa = 3.6ICEE185 pKa = 4.15IRR187 pKa = 11.84GKK189 pKa = 8.06TFEE192 pKa = 4.16VVTFVTSTEE201 pKa = 4.08GKK203 pKa = 8.91PVKK206 pKa = 10.36NLEE209 pKa = 4.37KK210 pKa = 10.58YY211 pKa = 7.39YY212 pKa = 10.71QCLTRR217 pKa = 11.84HH218 pKa = 5.03SSKK221 pKa = 10.99LKK223 pKa = 10.28ILSPDD228 pKa = 2.96ASFASSS234 pKa = 2.9
Molecular weight: 25.88 kDa
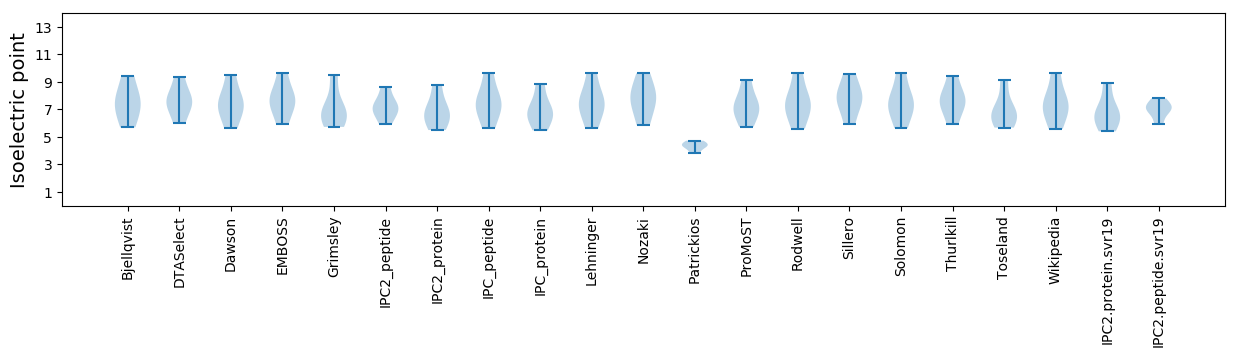
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R7Z6N1|A0A2R7Z6N1_9VIRU Helicase OS=Elm carlavirus OX=2015551 GN=replicase PE=3 SV=1
MM1 pKa = 7.65PLSPPADD8 pKa = 3.42KK9 pKa = 10.74TSVYY13 pKa = 9.54FASAIGLGIALSLFTLTRR31 pKa = 11.84STLPHH36 pKa = 6.64VGDD39 pKa = 4.96NIHH42 pKa = 6.4SLPHH46 pKa = 5.54GGRR49 pKa = 11.84YY50 pKa = 8.92RR51 pKa = 11.84DD52 pKa = 3.63GTKK55 pKa = 10.42SVDD58 pKa = 3.28YY59 pKa = 10.53CGPAKK64 pKa = 10.33RR65 pKa = 11.84YY66 pKa = 8.42PSSNLLTSGSSSTAVLFIIVLVGLIWLSDD95 pKa = 3.36KK96 pKa = 10.7FGKK99 pKa = 10.39RR100 pKa = 11.84GVCNNCNRR108 pKa = 11.84SHH110 pKa = 5.62QCC112 pKa = 3.38
MM1 pKa = 7.65PLSPPADD8 pKa = 3.42KK9 pKa = 10.74TSVYY13 pKa = 9.54FASAIGLGIALSLFTLTRR31 pKa = 11.84STLPHH36 pKa = 6.64VGDD39 pKa = 4.96NIHH42 pKa = 6.4SLPHH46 pKa = 5.54GGRR49 pKa = 11.84YY50 pKa = 8.92RR51 pKa = 11.84DD52 pKa = 3.63GTKK55 pKa = 10.42SVDD58 pKa = 3.28YY59 pKa = 10.53CGPAKK64 pKa = 10.33RR65 pKa = 11.84YY66 pKa = 8.42PSSNLLTSGSSSTAVLFIIVLVGLIWLSDD95 pKa = 3.36KK96 pKa = 10.7FGKK99 pKa = 10.39RR100 pKa = 11.84GVCNNCNRR108 pKa = 11.84SHH110 pKa = 5.62QCC112 pKa = 3.38
Molecular weight: 11.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2638 |
71 |
1915 |
527.6 |
59.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.255 ± 0.294 | 2.881 ± 0.37 |
5.648 ± 0.449 | 6.823 ± 0.982 |
5.951 ± 0.892 | 6.899 ± 0.578 |
2.578 ± 0.232 | 5.345 ± 0.501 |
7.771 ± 0.649 | 9.098 ± 0.851 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.414 | 4.094 ± 0.425 |
3.791 ± 0.578 | 2.35 ± 0.484 |
4.966 ± 0.296 | 7.24 ± 1.426 |
5.383 ± 0.635 | 6.52 ± 0.464 |
1.061 ± 0.197 | 2.881 ± 0.591 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
