
Fusarium graminearum mycovirus China 9
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Betachrysovirus; Fusarium graminearum chrysovirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
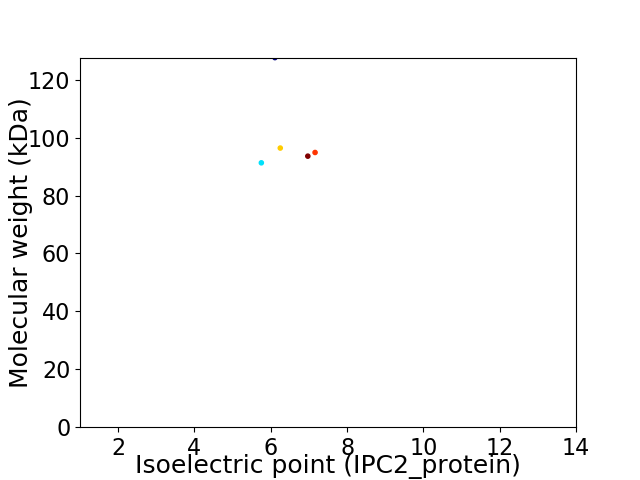
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|E7DDJ4|E7DDJ4_9VIRU Uncharacterized protein OS=Fusarium graminearum mycovirus China 9 OX=941336 PE=4 SV=2
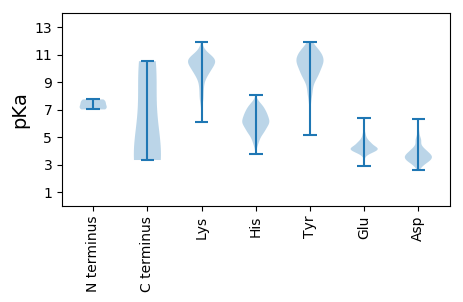
MM1 pKa = 7.07TTTFWKK7 pKa = 8.96STEE10 pKa = 4.26STDD13 pKa = 4.18SNSDD17 pKa = 3.34LAEE20 pKa = 3.7QRR22 pKa = 11.84AVHH25 pKa = 4.73STEE28 pKa = 3.93YY29 pKa = 10.57AVCTIHH35 pKa = 6.5QAWGLPSTPYY45 pKa = 10.94SLGGKK50 pKa = 8.04PDD52 pKa = 2.97QWEE55 pKa = 4.26FTTKK59 pKa = 10.84DD60 pKa = 3.39EE61 pKa = 4.76GSGPSKK67 pKa = 10.14LTGNVVDD74 pKa = 4.85GFGTLFSDD82 pKa = 4.75RR83 pKa = 11.84LFNLPSKK90 pKa = 10.91RR91 pKa = 11.84EE92 pKa = 3.83VTFFLHH98 pKa = 6.7SDD100 pKa = 3.29TAIEE104 pKa = 3.8MRR106 pKa = 11.84IINSNTFDD114 pKa = 3.93RR115 pKa = 11.84LEE117 pKa = 4.11NSCICVEE124 pKa = 3.99VCIVSRR130 pKa = 11.84ASPVQKK136 pKa = 10.07HH137 pKa = 5.37RR138 pKa = 11.84PVVLHH143 pKa = 5.81FWVYY147 pKa = 11.5DD148 pKa = 3.74EE149 pKa = 5.17FSGGQVKK156 pKa = 10.07IVLVEE161 pKa = 3.68TAYY164 pKa = 10.65DD165 pKa = 3.49YY166 pKa = 11.78SPFLRR171 pKa = 11.84IDD173 pKa = 3.18RR174 pKa = 11.84SGIGAAIRR182 pKa = 11.84SITSIHH188 pKa = 6.03TLGQTGTLNGSNVFSVVPWDD208 pKa = 3.68ANKK211 pKa = 10.35LQSCSVKK218 pKa = 10.26DD219 pKa = 3.56LRR221 pKa = 11.84RR222 pKa = 11.84LFVSTSNGKK231 pKa = 6.08TAKK234 pKa = 10.23LKK236 pKa = 10.91APLDD240 pKa = 3.56IKK242 pKa = 10.76QAAEE246 pKa = 4.21YY247 pKa = 9.68PLLIYY252 pKa = 10.75LGTSNDD258 pKa = 3.6HH259 pKa = 6.36EE260 pKa = 4.53NATRR264 pKa = 11.84MVLLRR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84KK272 pKa = 8.62DD273 pKa = 3.52TPDD276 pKa = 3.32TLCEE280 pKa = 3.9VLAQLLVRR288 pKa = 11.84ARR290 pKa = 11.84RR291 pKa = 11.84CPDD294 pKa = 2.95SQRR297 pKa = 11.84CDD299 pKa = 3.65DD300 pKa = 4.89ANDD303 pKa = 3.86PSAMLPSNVLVLATNGGKK321 pKa = 9.4QVTATCVAVRR331 pKa = 11.84VRR333 pKa = 11.84ADD335 pKa = 2.88GRR337 pKa = 11.84YY338 pKa = 9.44LYY340 pKa = 10.1STMRR344 pKa = 11.84RR345 pKa = 11.84NCLPYY350 pKa = 8.93TDD352 pKa = 3.84QVKK355 pKa = 8.38FTSIVPSAHH364 pKa = 5.58NLIMEE369 pKa = 4.58RR370 pKa = 11.84SFAVDD375 pKa = 2.57NWGSIMRR382 pKa = 11.84VNTNNSPVVVHH393 pKa = 5.83TAVTEE398 pKa = 4.38VIRR401 pKa = 11.84NAALGTTDD409 pKa = 2.85TTVYY413 pKa = 9.98VRR415 pKa = 11.84HH416 pKa = 5.92VLDD419 pKa = 4.41AFGTICSLMDD429 pKa = 3.23RR430 pKa = 11.84TVGLSYY436 pKa = 9.91EE437 pKa = 4.16IPQDD441 pKa = 3.84KK442 pKa = 10.41NFCQIRR448 pKa = 11.84WKK450 pKa = 10.7HH451 pKa = 6.05DD452 pKa = 3.42ALSLKK457 pKa = 9.04GTLFIASAVNPGTCLLGVLGSWQNWAGPGSGGGQIPQSQEE497 pKa = 2.91YY498 pKa = 8.56WYY500 pKa = 9.55PIDD503 pKa = 3.69GRR505 pKa = 11.84GGKK508 pKa = 9.27DD509 pKa = 2.95CYY511 pKa = 11.17GEE513 pKa = 5.11LMYY516 pKa = 10.97LDD518 pKa = 4.89GGFSLLGRR526 pKa = 11.84YY527 pKa = 9.1LYY529 pKa = 10.62QDD531 pKa = 4.34VINSGQYY538 pKa = 10.99LDD540 pKa = 3.93LTSTLVPKK548 pKa = 10.65ALRR551 pKa = 11.84DD552 pKa = 3.82DD553 pKa = 4.75GLITDD558 pKa = 4.01NLRR561 pKa = 11.84VYY563 pKa = 10.37VGPHH567 pKa = 6.27EE568 pKa = 4.35GDD570 pKa = 3.09KK571 pKa = 11.34VNFMLDD577 pKa = 2.86IEE579 pKa = 4.56YY580 pKa = 10.31TDD582 pKa = 5.02LPGGDD587 pKa = 3.99SKK589 pKa = 10.32TPTAIGYY596 pKa = 8.09CAVQQGEE603 pKa = 4.55SLSSGLSTMHH613 pKa = 6.57ATTLDD618 pKa = 3.79GYY620 pKa = 11.38GLITDD625 pKa = 4.75PKK627 pKa = 10.55SSAKK631 pKa = 10.14LDD633 pKa = 3.27QTLQQRR639 pKa = 11.84VGKK642 pKa = 9.58HH643 pKa = 4.22RR644 pKa = 11.84QLSKK648 pKa = 10.78RR649 pKa = 11.84YY650 pKa = 9.14ASSNLWEE657 pKa = 4.96DD658 pKa = 3.58LAVLQTLALSSNSVQLWAKK677 pKa = 10.3GADD680 pKa = 3.25NDD682 pKa = 4.81AAFLAAATPVNSSSCPTWNSIKK704 pKa = 10.64HH705 pKa = 5.44EE706 pKa = 4.26GAGLVANGTLLDD718 pKa = 4.16IGGEE722 pKa = 4.14LPKK725 pKa = 10.42FAALAKK731 pKa = 8.3RR732 pKa = 11.84HH733 pKa = 5.65KK734 pKa = 10.51VSSEE738 pKa = 3.77HH739 pKa = 6.0NPCIEE744 pKa = 4.05SALFAIEE751 pKa = 3.63AGIWRR756 pKa = 11.84KK757 pKa = 7.6TPDD760 pKa = 3.52LASVRR765 pKa = 11.84LSTTSLAEE773 pKa = 4.03GLADD777 pKa = 4.62LLNRR781 pKa = 11.84LYY783 pKa = 10.29TQDD786 pKa = 3.71VLIKK790 pKa = 10.62GDD792 pKa = 5.35DD793 pKa = 3.55GAEE796 pKa = 3.82WVEE799 pKa = 4.74GITGTFDD806 pKa = 2.85IHH808 pKa = 7.19VISPDD813 pKa = 3.15HH814 pKa = 6.86PARR817 pKa = 11.84EE818 pKa = 4.2ALRR821 pKa = 11.84IVTDD825 pKa = 3.73RR826 pKa = 11.84VIAASKK832 pKa = 10.65DD833 pKa = 3.8SNN835 pKa = 3.61
MM1 pKa = 7.07TTTFWKK7 pKa = 8.96STEE10 pKa = 4.26STDD13 pKa = 4.18SNSDD17 pKa = 3.34LAEE20 pKa = 3.7QRR22 pKa = 11.84AVHH25 pKa = 4.73STEE28 pKa = 3.93YY29 pKa = 10.57AVCTIHH35 pKa = 6.5QAWGLPSTPYY45 pKa = 10.94SLGGKK50 pKa = 8.04PDD52 pKa = 2.97QWEE55 pKa = 4.26FTTKK59 pKa = 10.84DD60 pKa = 3.39EE61 pKa = 4.76GSGPSKK67 pKa = 10.14LTGNVVDD74 pKa = 4.85GFGTLFSDD82 pKa = 4.75RR83 pKa = 11.84LFNLPSKK90 pKa = 10.91RR91 pKa = 11.84EE92 pKa = 3.83VTFFLHH98 pKa = 6.7SDD100 pKa = 3.29TAIEE104 pKa = 3.8MRR106 pKa = 11.84IINSNTFDD114 pKa = 3.93RR115 pKa = 11.84LEE117 pKa = 4.11NSCICVEE124 pKa = 3.99VCIVSRR130 pKa = 11.84ASPVQKK136 pKa = 10.07HH137 pKa = 5.37RR138 pKa = 11.84PVVLHH143 pKa = 5.81FWVYY147 pKa = 11.5DD148 pKa = 3.74EE149 pKa = 5.17FSGGQVKK156 pKa = 10.07IVLVEE161 pKa = 3.68TAYY164 pKa = 10.65DD165 pKa = 3.49YY166 pKa = 11.78SPFLRR171 pKa = 11.84IDD173 pKa = 3.18RR174 pKa = 11.84SGIGAAIRR182 pKa = 11.84SITSIHH188 pKa = 6.03TLGQTGTLNGSNVFSVVPWDD208 pKa = 3.68ANKK211 pKa = 10.35LQSCSVKK218 pKa = 10.26DD219 pKa = 3.56LRR221 pKa = 11.84RR222 pKa = 11.84LFVSTSNGKK231 pKa = 6.08TAKK234 pKa = 10.23LKK236 pKa = 10.91APLDD240 pKa = 3.56IKK242 pKa = 10.76QAAEE246 pKa = 4.21YY247 pKa = 9.68PLLIYY252 pKa = 10.75LGTSNDD258 pKa = 3.6HH259 pKa = 6.36EE260 pKa = 4.53NATRR264 pKa = 11.84MVLLRR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84KK272 pKa = 8.62DD273 pKa = 3.52TPDD276 pKa = 3.32TLCEE280 pKa = 3.9VLAQLLVRR288 pKa = 11.84ARR290 pKa = 11.84RR291 pKa = 11.84CPDD294 pKa = 2.95SQRR297 pKa = 11.84CDD299 pKa = 3.65DD300 pKa = 4.89ANDD303 pKa = 3.86PSAMLPSNVLVLATNGGKK321 pKa = 9.4QVTATCVAVRR331 pKa = 11.84VRR333 pKa = 11.84ADD335 pKa = 2.88GRR337 pKa = 11.84YY338 pKa = 9.44LYY340 pKa = 10.1STMRR344 pKa = 11.84RR345 pKa = 11.84NCLPYY350 pKa = 8.93TDD352 pKa = 3.84QVKK355 pKa = 8.38FTSIVPSAHH364 pKa = 5.58NLIMEE369 pKa = 4.58RR370 pKa = 11.84SFAVDD375 pKa = 2.57NWGSIMRR382 pKa = 11.84VNTNNSPVVVHH393 pKa = 5.83TAVTEE398 pKa = 4.38VIRR401 pKa = 11.84NAALGTTDD409 pKa = 2.85TTVYY413 pKa = 9.98VRR415 pKa = 11.84HH416 pKa = 5.92VLDD419 pKa = 4.41AFGTICSLMDD429 pKa = 3.23RR430 pKa = 11.84TVGLSYY436 pKa = 9.91EE437 pKa = 4.16IPQDD441 pKa = 3.84KK442 pKa = 10.41NFCQIRR448 pKa = 11.84WKK450 pKa = 10.7HH451 pKa = 6.05DD452 pKa = 3.42ALSLKK457 pKa = 9.04GTLFIASAVNPGTCLLGVLGSWQNWAGPGSGGGQIPQSQEE497 pKa = 2.91YY498 pKa = 8.56WYY500 pKa = 9.55PIDD503 pKa = 3.69GRR505 pKa = 11.84GGKK508 pKa = 9.27DD509 pKa = 2.95CYY511 pKa = 11.17GEE513 pKa = 5.11LMYY516 pKa = 10.97LDD518 pKa = 4.89GGFSLLGRR526 pKa = 11.84YY527 pKa = 9.1LYY529 pKa = 10.62QDD531 pKa = 4.34VINSGQYY538 pKa = 10.99LDD540 pKa = 3.93LTSTLVPKK548 pKa = 10.65ALRR551 pKa = 11.84DD552 pKa = 3.82DD553 pKa = 4.75GLITDD558 pKa = 4.01NLRR561 pKa = 11.84VYY563 pKa = 10.37VGPHH567 pKa = 6.27EE568 pKa = 4.35GDD570 pKa = 3.09KK571 pKa = 11.34VNFMLDD577 pKa = 2.86IEE579 pKa = 4.56YY580 pKa = 10.31TDD582 pKa = 5.02LPGGDD587 pKa = 3.99SKK589 pKa = 10.32TPTAIGYY596 pKa = 8.09CAVQQGEE603 pKa = 4.55SLSSGLSTMHH613 pKa = 6.57ATTLDD618 pKa = 3.79GYY620 pKa = 11.38GLITDD625 pKa = 4.75PKK627 pKa = 10.55SSAKK631 pKa = 10.14LDD633 pKa = 3.27QTLQQRR639 pKa = 11.84VGKK642 pKa = 9.58HH643 pKa = 4.22RR644 pKa = 11.84QLSKK648 pKa = 10.78RR649 pKa = 11.84YY650 pKa = 9.14ASSNLWEE657 pKa = 4.96DD658 pKa = 3.58LAVLQTLALSSNSVQLWAKK677 pKa = 10.3GADD680 pKa = 3.25NDD682 pKa = 4.81AAFLAAATPVNSSSCPTWNSIKK704 pKa = 10.64HH705 pKa = 5.44EE706 pKa = 4.26GAGLVANGTLLDD718 pKa = 4.16IGGEE722 pKa = 4.14LPKK725 pKa = 10.42FAALAKK731 pKa = 8.3RR732 pKa = 11.84HH733 pKa = 5.65KK734 pKa = 10.51VSSEE738 pKa = 3.77HH739 pKa = 6.0NPCIEE744 pKa = 4.05SALFAIEE751 pKa = 3.63AGIWRR756 pKa = 11.84KK757 pKa = 7.6TPDD760 pKa = 3.52LASVRR765 pKa = 11.84LSTTSLAEE773 pKa = 4.03GLADD777 pKa = 4.62LLNRR781 pKa = 11.84LYY783 pKa = 10.29TQDD786 pKa = 3.71VLIKK790 pKa = 10.62GDD792 pKa = 5.35DD793 pKa = 3.55GAEE796 pKa = 3.82WVEE799 pKa = 4.74GITGTFDD806 pKa = 2.85IHH808 pKa = 7.19VISPDD813 pKa = 3.15HH814 pKa = 6.86PARR817 pKa = 11.84EE818 pKa = 4.2ALRR821 pKa = 11.84IVTDD825 pKa = 3.73RR826 pKa = 11.84VIAASKK832 pKa = 10.65DD833 pKa = 3.8SNN835 pKa = 3.61
Molecular weight: 91.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7DDJ2|E7DDJ2_9VIRU Putative coat protein OS=Fusarium graminearum mycovirus China 9 OX=941336 PE=4 SV=1
MM1 pKa = 7.6SGQLNIRR8 pKa = 11.84STAHH12 pKa = 7.2LEE14 pKa = 4.16QQAVAPDD21 pKa = 3.56QYY23 pKa = 11.13RR24 pKa = 11.84AGAVAGLCSYY34 pKa = 10.38SVPTPSITRR43 pKa = 11.84RR44 pKa = 11.84VHH46 pKa = 5.9YY47 pKa = 9.83NAGVSTVEE55 pKa = 4.21SGAGSADD62 pKa = 3.31KK63 pKa = 11.29LEE65 pKa = 4.34SFYY68 pKa = 11.14HH69 pKa = 5.53HH70 pKa = 7.5RR71 pKa = 11.84IPRR74 pKa = 11.84DD75 pKa = 3.24VALTSLRR82 pKa = 11.84AGTIKK87 pKa = 10.52HH88 pKa = 6.39DD89 pKa = 3.7VVTFDD94 pKa = 3.12VSEE97 pKa = 4.6RR98 pKa = 11.84GPTQPLTNQIVNVTYY113 pKa = 9.56KK114 pKa = 9.89QQYY117 pKa = 8.21NFIDD121 pKa = 3.72VGVSAAIVRR130 pKa = 11.84TEE132 pKa = 3.8LQFFPVRR139 pKa = 11.84PPVDD143 pKa = 3.4NTKK146 pKa = 11.15LNLLKK151 pKa = 10.77NALTVHH157 pKa = 6.01EE158 pKa = 4.5VVKK161 pKa = 10.05PLKK164 pKa = 10.39LQLPKK169 pKa = 10.72LVAQSHH175 pKa = 6.69ASPGAQLYY183 pKa = 11.02SDD185 pKa = 4.59FEE187 pKa = 4.19PLRR190 pKa = 11.84RR191 pKa = 11.84AIANQAINDD200 pKa = 4.06DD201 pKa = 3.58YY202 pKa = 11.95GMRR205 pKa = 11.84LSHH208 pKa = 6.31MARR211 pKa = 11.84IVLHH215 pKa = 6.51SFTAQAAGLSHH226 pKa = 7.07AASDD230 pKa = 4.0VVSTVDD236 pKa = 3.14VRR238 pKa = 11.84TMFVAEE244 pKa = 4.98TYY246 pKa = 10.84QMSAAEE252 pKa = 4.0LQQRR256 pKa = 11.84AFVYY260 pKa = 10.69VDD262 pKa = 3.74DD263 pKa = 6.3DD264 pKa = 3.51ISEE267 pKa = 4.54EE268 pKa = 4.17YY269 pKa = 9.63WAFLAMAMLGINTNVGNTRR288 pKa = 11.84TVYY291 pKa = 10.51SRR293 pKa = 11.84LVSVSEE299 pKa = 4.07LRR301 pKa = 11.84GGQQLYY307 pKa = 10.02YY308 pKa = 10.97VMKK311 pKa = 10.56SGEE314 pKa = 4.13NTPANISQNACASVLNNPEE333 pKa = 4.03KK334 pKa = 10.9CLSFYY339 pKa = 10.91NAYY342 pKa = 9.96AQSLGLAQEE351 pKa = 4.25ASSVLRR357 pKa = 11.84QVAVMPFMFTKK368 pKa = 10.65NATLPYY374 pKa = 9.63KK375 pKa = 10.66ARR377 pKa = 11.84EE378 pKa = 4.03NPKK381 pKa = 10.04ADD383 pKa = 3.04AYY385 pKa = 11.25VYY387 pKa = 10.66LLRR390 pKa = 11.84EE391 pKa = 3.86TAEE394 pKa = 5.24DD395 pKa = 3.11IRR397 pKa = 11.84LMYY400 pKa = 10.73NSVEE404 pKa = 4.18TLVYY408 pKa = 9.06QAPIIAGAVKK418 pKa = 10.42AGVAAISMSYY428 pKa = 9.87RR429 pKa = 11.84VKK431 pKa = 11.03NSLNVPSILAAIKK444 pKa = 10.13EE445 pKa = 4.14ILADD449 pKa = 3.9PSSCRR454 pKa = 11.84STMSAVLCSYY464 pKa = 11.28GGITADD470 pKa = 5.46LEE472 pKa = 4.11WLLPFTEE479 pKa = 4.83GVDD482 pKa = 3.26IAMDD486 pKa = 3.69ACVRR490 pKa = 11.84AYY492 pKa = 10.33RR493 pKa = 11.84DD494 pKa = 4.09CGWLITTYY502 pKa = 11.4NDD504 pKa = 3.97LSCLGAYY511 pKa = 9.91APAFSTGVDD520 pKa = 3.2MRR522 pKa = 11.84NAQFGYY528 pKa = 10.67DD529 pKa = 3.62VASRR533 pKa = 11.84PYY535 pKa = 10.53LEE537 pKa = 4.79SLIALSTQGLPWTFEE552 pKa = 4.07NEE554 pKa = 4.35SLDD557 pKa = 3.88GPTLQRR563 pKa = 11.84AARR566 pKa = 11.84DD567 pKa = 3.86VVHH570 pKa = 6.6WEE572 pKa = 5.15AVIQWDD578 pKa = 4.14AYY580 pKa = 10.79SPGEE584 pKa = 4.28GVSVAHH590 pKa = 6.2ITPTPVSPKK599 pKa = 7.73QARR602 pKa = 11.84PFEE605 pKa = 4.45TEE607 pKa = 3.81PNAHH611 pKa = 6.98LSEE614 pKa = 4.5RR615 pKa = 11.84WQKK618 pKa = 10.53SKK620 pKa = 10.22RR621 pKa = 11.84TLSGNGLKK629 pKa = 10.51ALTLSPPSAAARR641 pKa = 11.84SEE643 pKa = 4.03ATKK646 pKa = 9.18TASVAEE652 pKa = 4.23TLRR655 pKa = 11.84THH657 pKa = 5.38TRR659 pKa = 11.84SMSVKK664 pKa = 9.59PNPAPGTKK672 pKa = 8.63PHH674 pKa = 6.01NMPWPVQAAASVPSGEE690 pKa = 4.39VSDD693 pKa = 4.97SEE695 pKa = 4.6STTTRR700 pKa = 11.84VTRR703 pKa = 11.84ANSVASTDD711 pKa = 3.81DD712 pKa = 3.34TASAATVVQTAPPGVTSPPRR732 pKa = 11.84GRR734 pKa = 11.84SVTPIGTTVDD744 pKa = 3.93LSRR747 pKa = 11.84EE748 pKa = 4.19PTPSLAEE755 pKa = 3.88RR756 pKa = 11.84LKK758 pKa = 11.19SPFRR762 pKa = 11.84HH763 pKa = 4.36QRR765 pKa = 11.84VRR767 pKa = 11.84SRR769 pKa = 11.84SVSSSRR775 pKa = 11.84GRR777 pKa = 11.84ATSDD781 pKa = 2.84TNMGQMLDD789 pKa = 3.47RR790 pKa = 11.84GASEE794 pKa = 4.01SSEE797 pKa = 3.45IALIRR802 pKa = 11.84KK803 pKa = 7.0TKK805 pKa = 9.87PKK807 pKa = 10.5DD808 pKa = 3.37VAEE811 pKa = 4.48LGEE814 pKa = 4.87HH815 pKa = 6.33IVTSDD820 pKa = 3.17GRR822 pKa = 11.84IEE824 pKa = 6.35LITQLAGKK832 pKa = 9.54RR833 pKa = 11.84DD834 pKa = 3.76YY835 pKa = 11.65SDD837 pKa = 3.29VVRR840 pKa = 11.84DD841 pKa = 3.95LLPGGVGTPDD851 pKa = 2.95VGDD854 pKa = 3.29WTGTSKK860 pKa = 10.96NGEE863 pKa = 3.91QRR865 pKa = 11.84NITTLMNVNKK875 pKa = 10.52
MM1 pKa = 7.6SGQLNIRR8 pKa = 11.84STAHH12 pKa = 7.2LEE14 pKa = 4.16QQAVAPDD21 pKa = 3.56QYY23 pKa = 11.13RR24 pKa = 11.84AGAVAGLCSYY34 pKa = 10.38SVPTPSITRR43 pKa = 11.84RR44 pKa = 11.84VHH46 pKa = 5.9YY47 pKa = 9.83NAGVSTVEE55 pKa = 4.21SGAGSADD62 pKa = 3.31KK63 pKa = 11.29LEE65 pKa = 4.34SFYY68 pKa = 11.14HH69 pKa = 5.53HH70 pKa = 7.5RR71 pKa = 11.84IPRR74 pKa = 11.84DD75 pKa = 3.24VALTSLRR82 pKa = 11.84AGTIKK87 pKa = 10.52HH88 pKa = 6.39DD89 pKa = 3.7VVTFDD94 pKa = 3.12VSEE97 pKa = 4.6RR98 pKa = 11.84GPTQPLTNQIVNVTYY113 pKa = 9.56KK114 pKa = 9.89QQYY117 pKa = 8.21NFIDD121 pKa = 3.72VGVSAAIVRR130 pKa = 11.84TEE132 pKa = 3.8LQFFPVRR139 pKa = 11.84PPVDD143 pKa = 3.4NTKK146 pKa = 11.15LNLLKK151 pKa = 10.77NALTVHH157 pKa = 6.01EE158 pKa = 4.5VVKK161 pKa = 10.05PLKK164 pKa = 10.39LQLPKK169 pKa = 10.72LVAQSHH175 pKa = 6.69ASPGAQLYY183 pKa = 11.02SDD185 pKa = 4.59FEE187 pKa = 4.19PLRR190 pKa = 11.84RR191 pKa = 11.84AIANQAINDD200 pKa = 4.06DD201 pKa = 3.58YY202 pKa = 11.95GMRR205 pKa = 11.84LSHH208 pKa = 6.31MARR211 pKa = 11.84IVLHH215 pKa = 6.51SFTAQAAGLSHH226 pKa = 7.07AASDD230 pKa = 4.0VVSTVDD236 pKa = 3.14VRR238 pKa = 11.84TMFVAEE244 pKa = 4.98TYY246 pKa = 10.84QMSAAEE252 pKa = 4.0LQQRR256 pKa = 11.84AFVYY260 pKa = 10.69VDD262 pKa = 3.74DD263 pKa = 6.3DD264 pKa = 3.51ISEE267 pKa = 4.54EE268 pKa = 4.17YY269 pKa = 9.63WAFLAMAMLGINTNVGNTRR288 pKa = 11.84TVYY291 pKa = 10.51SRR293 pKa = 11.84LVSVSEE299 pKa = 4.07LRR301 pKa = 11.84GGQQLYY307 pKa = 10.02YY308 pKa = 10.97VMKK311 pKa = 10.56SGEE314 pKa = 4.13NTPANISQNACASVLNNPEE333 pKa = 4.03KK334 pKa = 10.9CLSFYY339 pKa = 10.91NAYY342 pKa = 9.96AQSLGLAQEE351 pKa = 4.25ASSVLRR357 pKa = 11.84QVAVMPFMFTKK368 pKa = 10.65NATLPYY374 pKa = 9.63KK375 pKa = 10.66ARR377 pKa = 11.84EE378 pKa = 4.03NPKK381 pKa = 10.04ADD383 pKa = 3.04AYY385 pKa = 11.25VYY387 pKa = 10.66LLRR390 pKa = 11.84EE391 pKa = 3.86TAEE394 pKa = 5.24DD395 pKa = 3.11IRR397 pKa = 11.84LMYY400 pKa = 10.73NSVEE404 pKa = 4.18TLVYY408 pKa = 9.06QAPIIAGAVKK418 pKa = 10.42AGVAAISMSYY428 pKa = 9.87RR429 pKa = 11.84VKK431 pKa = 11.03NSLNVPSILAAIKK444 pKa = 10.13EE445 pKa = 4.14ILADD449 pKa = 3.9PSSCRR454 pKa = 11.84STMSAVLCSYY464 pKa = 11.28GGITADD470 pKa = 5.46LEE472 pKa = 4.11WLLPFTEE479 pKa = 4.83GVDD482 pKa = 3.26IAMDD486 pKa = 3.69ACVRR490 pKa = 11.84AYY492 pKa = 10.33RR493 pKa = 11.84DD494 pKa = 4.09CGWLITTYY502 pKa = 11.4NDD504 pKa = 3.97LSCLGAYY511 pKa = 9.91APAFSTGVDD520 pKa = 3.2MRR522 pKa = 11.84NAQFGYY528 pKa = 10.67DD529 pKa = 3.62VASRR533 pKa = 11.84PYY535 pKa = 10.53LEE537 pKa = 4.79SLIALSTQGLPWTFEE552 pKa = 4.07NEE554 pKa = 4.35SLDD557 pKa = 3.88GPTLQRR563 pKa = 11.84AARR566 pKa = 11.84DD567 pKa = 3.86VVHH570 pKa = 6.6WEE572 pKa = 5.15AVIQWDD578 pKa = 4.14AYY580 pKa = 10.79SPGEE584 pKa = 4.28GVSVAHH590 pKa = 6.2ITPTPVSPKK599 pKa = 7.73QARR602 pKa = 11.84PFEE605 pKa = 4.45TEE607 pKa = 3.81PNAHH611 pKa = 6.98LSEE614 pKa = 4.5RR615 pKa = 11.84WQKK618 pKa = 10.53SKK620 pKa = 10.22RR621 pKa = 11.84TLSGNGLKK629 pKa = 10.51ALTLSPPSAAARR641 pKa = 11.84SEE643 pKa = 4.03ATKK646 pKa = 9.18TASVAEE652 pKa = 4.23TLRR655 pKa = 11.84THH657 pKa = 5.38TRR659 pKa = 11.84SMSVKK664 pKa = 9.59PNPAPGTKK672 pKa = 8.63PHH674 pKa = 6.01NMPWPVQAAASVPSGEE690 pKa = 4.39VSDD693 pKa = 4.97SEE695 pKa = 4.6STTTRR700 pKa = 11.84VTRR703 pKa = 11.84ANSVASTDD711 pKa = 3.81DD712 pKa = 3.34TASAATVVQTAPPGVTSPPRR732 pKa = 11.84GRR734 pKa = 11.84SVTPIGTTVDD744 pKa = 3.93LSRR747 pKa = 11.84EE748 pKa = 4.19PTPSLAEE755 pKa = 3.88RR756 pKa = 11.84LKK758 pKa = 11.19SPFRR762 pKa = 11.84HH763 pKa = 4.36QRR765 pKa = 11.84VRR767 pKa = 11.84SRR769 pKa = 11.84SVSSSRR775 pKa = 11.84GRR777 pKa = 11.84ATSDD781 pKa = 2.84TNMGQMLDD789 pKa = 3.47RR790 pKa = 11.84GASEE794 pKa = 4.01SSEE797 pKa = 3.45IALIRR802 pKa = 11.84KK803 pKa = 7.0TKK805 pKa = 9.87PKK807 pKa = 10.5DD808 pKa = 3.37VAEE811 pKa = 4.48LGEE814 pKa = 4.87HH815 pKa = 6.33IVTSDD820 pKa = 3.17GRR822 pKa = 11.84IEE824 pKa = 6.35LITQLAGKK832 pKa = 9.54RR833 pKa = 11.84DD834 pKa = 3.76YY835 pKa = 11.65SDD837 pKa = 3.29VVRR840 pKa = 11.84DD841 pKa = 3.95LLPGGVGTPDD851 pKa = 2.95VGDD854 pKa = 3.29WTGTSKK860 pKa = 10.96NGEE863 pKa = 3.91QRR865 pKa = 11.84NITTLMNVNKK875 pKa = 10.52
Molecular weight: 94.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4556 |
835 |
1137 |
911.2 |
100.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.206 ± 0.559 | 2.019 ± 0.68 |
5.948 ± 0.366 | 5.114 ± 0.472 |
2.788 ± 0.212 | 6.255 ± 0.676 |
3.622 ± 1.152 | 3.512 ± 0.4 |
3.775 ± 0.342 | 8.275 ± 0.457 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.195 ± 0.33 | 3.929 ± 0.275 |
4.587 ± 0.434 | 4.368 ± 0.372 |
6.738 ± 0.383 | 8.033 ± 0.541 |
6.497 ± 0.421 | 7.309 ± 0.599 |
1.449 ± 0.146 | 3.38 ± 0.268 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
