
Striped jack nervous necrosis virus (SjNNV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Magsaviricetes; Nodamuvirales; Nodaviridae; Betanodavirus
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
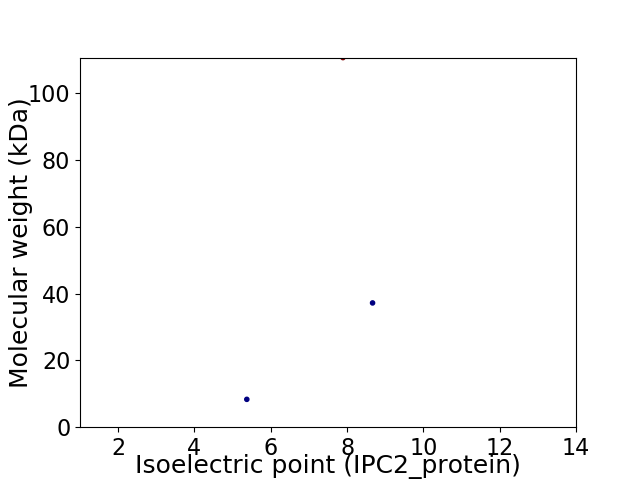
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
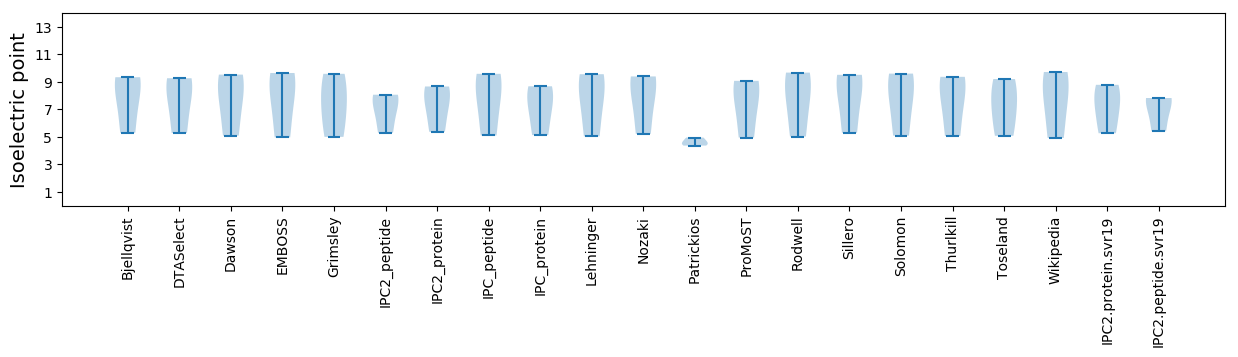
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9QAZ8|RDRP_SJNNV RNA-directed RNA polymerase OS=Striped jack nervous necrosis virus OX=35297 PE=3 SV=1
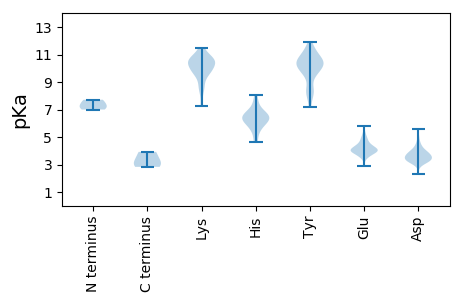
MM1 pKa = 7.29EE2 pKa = 4.59QVQQAIDD9 pKa = 3.29QHH11 pKa = 7.44LVEE14 pKa = 5.16LEE16 pKa = 4.05QLFKK20 pKa = 10.76MLMDD24 pKa = 3.29VRR26 pKa = 11.84VALGGVTVVQVNEE39 pKa = 3.64MRR41 pKa = 11.84TFVISASAAAQRR53 pKa = 11.84LRR55 pKa = 11.84ALARR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 7.53PAPLAVAAEE70 pKa = 4.09PMEE73 pKa = 4.49TEE75 pKa = 3.92
MM1 pKa = 7.29EE2 pKa = 4.59QVQQAIDD9 pKa = 3.29QHH11 pKa = 7.44LVEE14 pKa = 5.16LEE16 pKa = 4.05QLFKK20 pKa = 10.76MLMDD24 pKa = 3.29VRR26 pKa = 11.84VALGGVTVVQVNEE39 pKa = 3.64MRR41 pKa = 11.84TFVISASAAAQRR53 pKa = 11.84LRR55 pKa = 11.84ALARR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 7.53PAPLAVAAEE70 pKa = 4.09PMEE73 pKa = 4.49TEE75 pKa = 3.92
Molecular weight: 8.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9QAZ7|B_SJNNV Protein B OS=Striped jack nervous necrosis virus OX=35297 PE=4 SV=1
MM1 pKa = 7.0VRR3 pKa = 11.84KK4 pKa = 9.73GDD6 pKa = 3.65KK7 pKa = 10.35KK8 pKa = 10.41LAKK11 pKa = 10.21PPTTKK16 pKa = 10.51AANSQPRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84ATQRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SGRR34 pKa = 11.84ADD36 pKa = 3.27APLAKK41 pKa = 10.21ASTITGFGRR50 pKa = 11.84ATNDD54 pKa = 2.97VHH56 pKa = 8.04ISGMSRR62 pKa = 11.84IAQAVVPAGTGTDD75 pKa = 3.25GKK77 pKa = 10.48IVVDD81 pKa = 3.61STIVPEE87 pKa = 3.99LLPRR91 pKa = 11.84LGHH94 pKa = 6.87AARR97 pKa = 11.84IFQRR101 pKa = 11.84YY102 pKa = 8.09AVEE105 pKa = 3.9TLEE108 pKa = 5.81FEE110 pKa = 4.34IQPMCPANTGGGYY123 pKa = 9.72VAGFLPDD130 pKa = 3.64PTDD133 pKa = 3.5NDD135 pKa = 3.63HH136 pKa = 7.11TFDD139 pKa = 4.08ALQATRR145 pKa = 11.84GAVVAKK151 pKa = 8.41WWEE154 pKa = 4.1SRR156 pKa = 11.84TVRR159 pKa = 11.84PQYY162 pKa = 10.64TRR164 pKa = 11.84TLLWTSTGKK173 pKa = 8.32EE174 pKa = 3.75QRR176 pKa = 11.84LTSPGRR182 pKa = 11.84LVLLCVGSNTDD193 pKa = 3.35VVNVSVMCRR202 pKa = 11.84WSVRR206 pKa = 11.84LSVPSLEE213 pKa = 4.4TPEE216 pKa = 4.81DD217 pKa = 3.34TTAPITTQAPLHH229 pKa = 6.35NDD231 pKa = 3.79SINNGYY237 pKa = 8.18TGFRR241 pKa = 11.84SILLGATQLDD251 pKa = 4.44LAPANAVFVTDD262 pKa = 3.98KK263 pKa = 10.63PLPIDD268 pKa = 3.6YY269 pKa = 10.36NLGVGDD275 pKa = 3.89VDD277 pKa = 4.33RR278 pKa = 11.84AVYY281 pKa = 8.48WHH283 pKa = 6.63LRR285 pKa = 11.84KK286 pKa = 9.72KK287 pKa = 10.78AGDD290 pKa = 3.66TQVPAGYY297 pKa = 9.9FDD299 pKa = 3.22WGLWDD304 pKa = 4.95DD305 pKa = 4.07FNKK308 pKa = 9.77TFTVGAPYY316 pKa = 10.82YY317 pKa = 10.5SDD319 pKa = 3.28QQPRR323 pKa = 11.84QILLPAGTLFTRR335 pKa = 11.84VDD337 pKa = 3.75SEE339 pKa = 4.19NN340 pKa = 3.16
MM1 pKa = 7.0VRR3 pKa = 11.84KK4 pKa = 9.73GDD6 pKa = 3.65KK7 pKa = 10.35KK8 pKa = 10.41LAKK11 pKa = 10.21PPTTKK16 pKa = 10.51AANSQPRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84ATQRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SGRR34 pKa = 11.84ADD36 pKa = 3.27APLAKK41 pKa = 10.21ASTITGFGRR50 pKa = 11.84ATNDD54 pKa = 2.97VHH56 pKa = 8.04ISGMSRR62 pKa = 11.84IAQAVVPAGTGTDD75 pKa = 3.25GKK77 pKa = 10.48IVVDD81 pKa = 3.61STIVPEE87 pKa = 3.99LLPRR91 pKa = 11.84LGHH94 pKa = 6.87AARR97 pKa = 11.84IFQRR101 pKa = 11.84YY102 pKa = 8.09AVEE105 pKa = 3.9TLEE108 pKa = 5.81FEE110 pKa = 4.34IQPMCPANTGGGYY123 pKa = 9.72VAGFLPDD130 pKa = 3.64PTDD133 pKa = 3.5NDD135 pKa = 3.63HH136 pKa = 7.11TFDD139 pKa = 4.08ALQATRR145 pKa = 11.84GAVVAKK151 pKa = 8.41WWEE154 pKa = 4.1SRR156 pKa = 11.84TVRR159 pKa = 11.84PQYY162 pKa = 10.64TRR164 pKa = 11.84TLLWTSTGKK173 pKa = 8.32EE174 pKa = 3.75QRR176 pKa = 11.84LTSPGRR182 pKa = 11.84LVLLCVGSNTDD193 pKa = 3.35VVNVSVMCRR202 pKa = 11.84WSVRR206 pKa = 11.84LSVPSLEE213 pKa = 4.4TPEE216 pKa = 4.81DD217 pKa = 3.34TTAPITTQAPLHH229 pKa = 6.35NDD231 pKa = 3.79SINNGYY237 pKa = 8.18TGFRR241 pKa = 11.84SILLGATQLDD251 pKa = 4.44LAPANAVFVTDD262 pKa = 3.98KK263 pKa = 10.63PLPIDD268 pKa = 3.6YY269 pKa = 10.36NLGVGDD275 pKa = 3.89VDD277 pKa = 4.33RR278 pKa = 11.84AVYY281 pKa = 8.48WHH283 pKa = 6.63LRR285 pKa = 11.84KK286 pKa = 9.72KK287 pKa = 10.78AGDD290 pKa = 3.66TQVPAGYY297 pKa = 9.9FDD299 pKa = 3.22WGLWDD304 pKa = 4.95DD305 pKa = 4.07FNKK308 pKa = 9.77TFTVGAPYY316 pKa = 10.82YY317 pKa = 10.5SDD319 pKa = 3.28QQPRR323 pKa = 11.84QILLPAGTLFTRR335 pKa = 11.84VDD337 pKa = 3.75SEE339 pKa = 4.19NN340 pKa = 3.16
Molecular weight: 37.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1398 |
75 |
983 |
466.0 |
52.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.013 ± 1.017 | 1.216 ± 0.251 |
6.652 ± 0.524 | 4.578 ± 0.832 |
3.076 ± 0.07 | 6.438 ± 0.652 |
2.289 ± 0.406 | 3.934 ± 0.28 |
3.934 ± 0.371 | 8.155 ± 0.353 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.717 ± 0.626 | 3.505 ± 0.276 |
5.651 ± 0.548 | 4.649 ± 0.592 |
8.369 ± 0.322 | 6.652 ± 0.911 |
7.368 ± 1.027 | 7.582 ± 0.912 |
1.431 ± 0.264 | 3.791 ± 0.672 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
