
Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides thetaiotaomicron
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
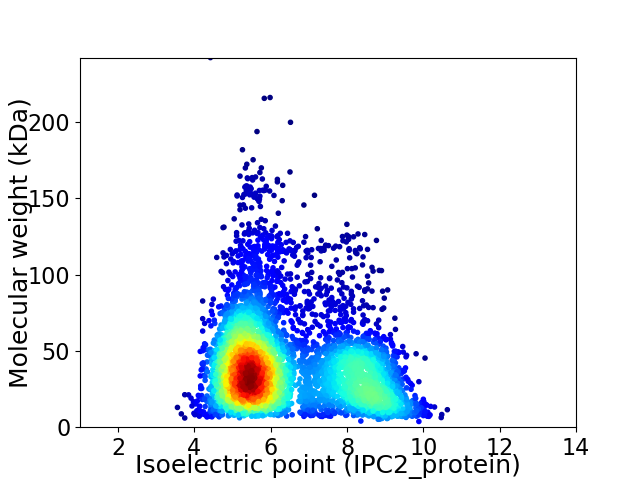
Virtual 2D-PAGE plot for 4782 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q89Z02|Q89Z02_BACTN Uncharacterized protein OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482) OX=226186 GN=BT_4575 PE=4 SV=1
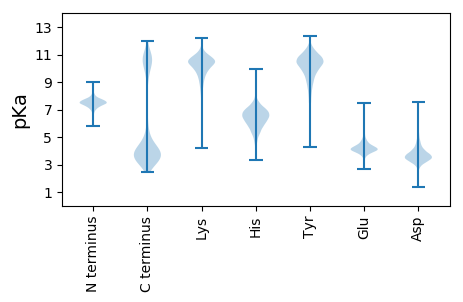
MM1 pKa = 7.58LKK3 pKa = 10.47FKK5 pKa = 10.66FGAWAVVLMLTAFSFSACDD24 pKa = 3.9DD25 pKa = 4.25NDD27 pKa = 4.11DD28 pKa = 3.73EE29 pKa = 5.27TYY31 pKa = 11.07NPPANITEE39 pKa = 4.32ALKK42 pKa = 10.5QLYY45 pKa = 9.8PNAQNVEE52 pKa = 3.91WEE54 pKa = 4.36MKK56 pKa = 9.77GDD58 pKa = 4.08YY59 pKa = 10.28YY60 pKa = 11.67VADD63 pKa = 3.66CWVTGDD69 pKa = 4.82EE70 pKa = 4.31LDD72 pKa = 3.44VWFDD76 pKa = 4.11ANANWVMTEE85 pKa = 4.13NEE87 pKa = 4.74LDD89 pKa = 4.42SIDD92 pKa = 3.83QLVPAVYY99 pKa = 9.68TGFRR103 pKa = 11.84NSNYY107 pKa = 9.88SSWVVTDD114 pKa = 3.72VFVLTYY120 pKa = 8.74PQHH123 pKa = 5.61PTEE126 pKa = 4.56SVIQVKK132 pKa = 10.11QGNLRR137 pKa = 11.84FALYY141 pKa = 10.07FSQEE145 pKa = 3.42GGLLHH150 pKa = 6.93EE151 pKa = 5.02RR152 pKa = 11.84DD153 pKa = 3.54ITNGDD158 pKa = 3.49DD159 pKa = 3.86TNWPVMEE166 pKa = 4.47
MM1 pKa = 7.58LKK3 pKa = 10.47FKK5 pKa = 10.66FGAWAVVLMLTAFSFSACDD24 pKa = 3.9DD25 pKa = 4.25NDD27 pKa = 4.11DD28 pKa = 3.73EE29 pKa = 5.27TYY31 pKa = 11.07NPPANITEE39 pKa = 4.32ALKK42 pKa = 10.5QLYY45 pKa = 9.8PNAQNVEE52 pKa = 3.91WEE54 pKa = 4.36MKK56 pKa = 9.77GDD58 pKa = 4.08YY59 pKa = 10.28YY60 pKa = 11.67VADD63 pKa = 3.66CWVTGDD69 pKa = 4.82EE70 pKa = 4.31LDD72 pKa = 3.44VWFDD76 pKa = 4.11ANANWVMTEE85 pKa = 4.13NEE87 pKa = 4.74LDD89 pKa = 4.42SIDD92 pKa = 3.83QLVPAVYY99 pKa = 9.68TGFRR103 pKa = 11.84NSNYY107 pKa = 9.88SSWVVTDD114 pKa = 3.72VFVLTYY120 pKa = 8.74PQHH123 pKa = 5.61PTEE126 pKa = 4.56SVIQVKK132 pKa = 10.11QGNLRR137 pKa = 11.84FALYY141 pKa = 10.07FSQEE145 pKa = 3.42GGLLHH150 pKa = 6.93EE151 pKa = 5.02RR152 pKa = 11.84DD153 pKa = 3.54ITNGDD158 pKa = 3.49DD159 pKa = 3.86TNWPVMEE166 pKa = 4.47
Molecular weight: 19.03 kDa
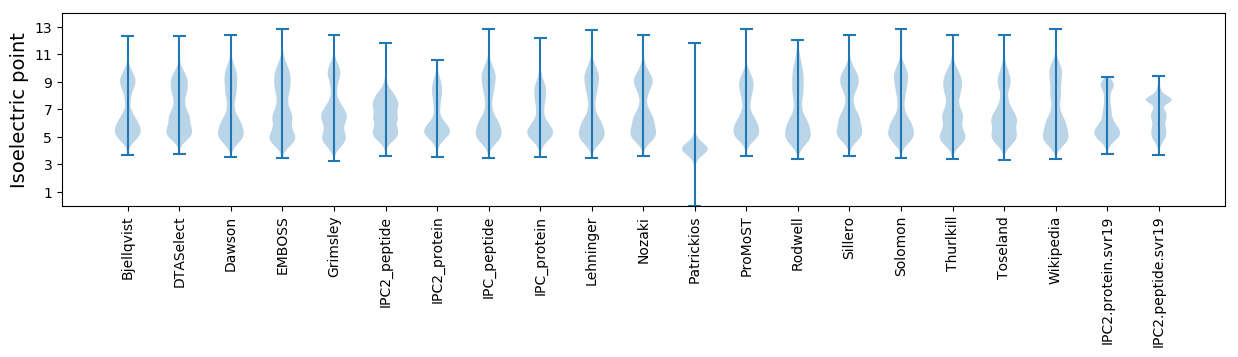
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8A5S5|RS6_BACTN 30S ribosomal protein S6 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482) OX=226186 GN=rpsF PE=3 SV=1
MM1 pKa = 7.62ARR3 pKa = 11.84NCRR6 pKa = 11.84NSSVSLTTYY15 pKa = 10.41SIVARR20 pKa = 11.84SVRR23 pKa = 11.84ITLNSSLKK31 pKa = 10.01IRR33 pKa = 11.84AAMNLDD39 pKa = 3.52LDD41 pKa = 4.15NRR43 pKa = 11.84PDD45 pKa = 3.61CSNMVRR51 pKa = 11.84NRR53 pKa = 11.84AWPASSRR60 pKa = 11.84LTVSRR65 pKa = 11.84RR66 pKa = 11.84MAGFGLGNLPPFPRR80 pKa = 11.84VRR82 pKa = 11.84GRR84 pKa = 11.84FFSLFIYY91 pKa = 9.07TFFMWYY97 pKa = 10.6NSTGWNCLRR106 pKa = 11.84QLPLPQFATSEE117 pKa = 4.1QIAAPLQGQAFFAAKK132 pKa = 10.16RR133 pKa = 11.84SGAAKK138 pKa = 8.85NTACYY143 pKa = 10.57VLANINLSSLEE154 pKa = 3.83RR155 pKa = 11.84KK156 pKa = 9.07EE157 pKa = 3.81RR158 pKa = 11.84FRR160 pKa = 11.84GKK162 pKa = 10.25RR163 pKa = 11.84GLVTDD168 pKa = 4.74GRR170 pKa = 11.84QTAHH174 pKa = 6.37SRR176 pKa = 11.84LDD178 pKa = 3.6GAKK181 pKa = 9.82LGSDD185 pKa = 4.01IISCISLTLATLCQMEE201 pKa = 4.62ATCAHH206 pKa = 6.11GWNRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 10.3NGRR215 pKa = 11.84PMMAEE220 pKa = 3.91SAVEE224 pKa = 5.52DD225 pKa = 3.95VPEE228 pKa = 3.93WDD230 pKa = 3.3AA231 pKa = 5.11
MM1 pKa = 7.62ARR3 pKa = 11.84NCRR6 pKa = 11.84NSSVSLTTYY15 pKa = 10.41SIVARR20 pKa = 11.84SVRR23 pKa = 11.84ITLNSSLKK31 pKa = 10.01IRR33 pKa = 11.84AAMNLDD39 pKa = 3.52LDD41 pKa = 4.15NRR43 pKa = 11.84PDD45 pKa = 3.61CSNMVRR51 pKa = 11.84NRR53 pKa = 11.84AWPASSRR60 pKa = 11.84LTVSRR65 pKa = 11.84RR66 pKa = 11.84MAGFGLGNLPPFPRR80 pKa = 11.84VRR82 pKa = 11.84GRR84 pKa = 11.84FFSLFIYY91 pKa = 9.07TFFMWYY97 pKa = 10.6NSTGWNCLRR106 pKa = 11.84QLPLPQFATSEE117 pKa = 4.1QIAAPLQGQAFFAAKK132 pKa = 10.16RR133 pKa = 11.84SGAAKK138 pKa = 8.85NTACYY143 pKa = 10.57VLANINLSSLEE154 pKa = 3.83RR155 pKa = 11.84KK156 pKa = 9.07EE157 pKa = 3.81RR158 pKa = 11.84FRR160 pKa = 11.84GKK162 pKa = 10.25RR163 pKa = 11.84GLVTDD168 pKa = 4.74GRR170 pKa = 11.84QTAHH174 pKa = 6.37SRR176 pKa = 11.84LDD178 pKa = 3.6GAKK181 pKa = 9.82LGSDD185 pKa = 4.01IISCISLTLATLCQMEE201 pKa = 4.62ATCAHH206 pKa = 6.11GWNRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 10.3NGRR215 pKa = 11.84PMMAEE220 pKa = 3.91SAVEE224 pKa = 5.52DD225 pKa = 3.95VPEE228 pKa = 3.93WDD230 pKa = 3.3AA231 pKa = 5.11
Molecular weight: 25.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1864427 |
32 |
2183 |
389.9 |
44.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.842 ± 0.032 | 1.22 ± 0.013 |
5.54 ± 0.023 | 6.516 ± 0.031 |
4.57 ± 0.021 | 6.78 ± 0.032 |
1.815 ± 0.014 | 6.889 ± 0.028 |
6.68 ± 0.025 | 9.016 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.717 ± 0.013 | 5.259 ± 0.029 |
3.784 ± 0.019 | 3.504 ± 0.018 |
4.546 ± 0.023 | 6.297 ± 0.026 |
5.709 ± 0.026 | 6.295 ± 0.026 |
1.365 ± 0.014 | 4.656 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
