
Panicum streak virus - Karino
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus; Panicum streak virus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
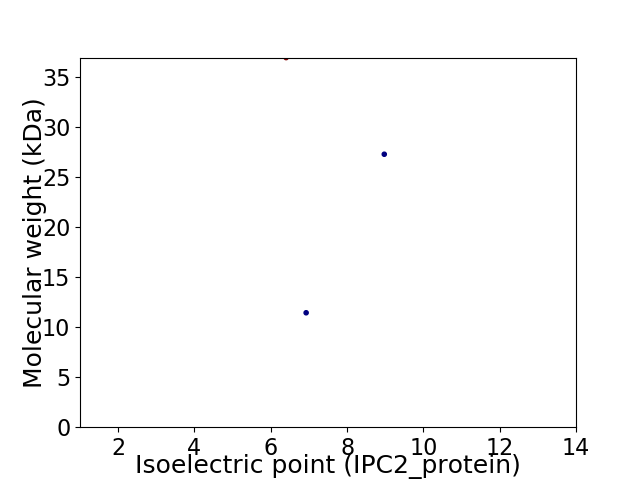
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q84370|Q84370_9GEMI Replication-associated protein OS=Panicum streak virus - Karino OX=268779 GN=RepA PE=3 SV=1
MM1 pKa = 6.97STSLSITSDD10 pKa = 2.59GRR12 pKa = 11.84HH13 pKa = 4.58SVRR16 pKa = 11.84SFRR19 pKa = 11.84HH20 pKa = 5.12RR21 pKa = 11.84NANTFLTYY29 pKa = 10.26SKK31 pKa = 10.88CPLEE35 pKa = 3.93PEE37 pKa = 4.86FIGEE41 pKa = 3.97HH42 pKa = 6.35LFRR45 pKa = 11.84LTKK48 pKa = 10.5DD49 pKa = 3.58FEE51 pKa = 4.03PAYY54 pKa = 9.19ILVVRR59 pKa = 11.84EE60 pKa = 3.81THH62 pKa = 6.89QDD64 pKa = 3.85GTWHH68 pKa = 6.13CHH70 pKa = 6.66ALLQCIKK77 pKa = 10.37PVTTRR82 pKa = 11.84DD83 pKa = 3.08EE84 pKa = 4.6RR85 pKa = 11.84YY86 pKa = 9.54FDD88 pKa = 3.44IDD90 pKa = 3.67RR91 pKa = 11.84YY92 pKa = 10.35HH93 pKa = 7.33PNIQSAKK100 pKa = 8.17STDD103 pKa = 3.09KK104 pKa = 10.55VRR106 pKa = 11.84EE107 pKa = 4.1YY108 pKa = 10.68ILKK111 pKa = 10.31DD112 pKa = 3.71PKK114 pKa = 11.09DD115 pKa = 3.29KK116 pKa = 10.5WEE118 pKa = 4.22KK119 pKa = 8.82GTYY122 pKa = 9.25IPRR125 pKa = 11.84KK126 pKa = 9.51KK127 pKa = 10.78SFVPPGKK134 pKa = 10.11EE135 pKa = 3.6NSEE138 pKa = 4.16KK139 pKa = 10.64KK140 pKa = 9.44PSKK143 pKa = 10.63DD144 pKa = 3.13EE145 pKa = 3.95VMKK148 pKa = 10.81EE149 pKa = 3.71IMTHH153 pKa = 4.37ATSRR157 pKa = 11.84AEE159 pKa = 3.89YY160 pKa = 10.09LSLVQTSLPYY170 pKa = 10.68DD171 pKa = 3.35WATKK175 pKa = 10.42LSYY178 pKa = 10.7FEE180 pKa = 4.77YY181 pKa = 10.06SASRR185 pKa = 11.84LFPDD189 pKa = 3.14IAEE192 pKa = 4.97PYY194 pKa = 9.71SNPHH198 pKa = 6.94PATDD202 pKa = 4.42PDD204 pKa = 4.21LLCNEE209 pKa = 4.77TLQDD213 pKa = 3.25WLEE216 pKa = 3.89PNIYY220 pKa = 9.6QVSPKK225 pKa = 10.59AYY227 pKa = 8.25MLLEE231 pKa = 4.21PSCLSLEE238 pKa = 4.12QAKK241 pKa = 10.7ADD243 pKa = 4.11LEE245 pKa = 4.43WLSEE249 pKa = 4.2TTRR252 pKa = 11.84LFQEE256 pKa = 4.37QEE258 pKa = 4.35SEE260 pKa = 4.12ASTSSAQHH268 pKa = 5.76GQVKK272 pKa = 10.26HH273 pKa = 6.34PGPEE277 pKa = 4.01ASDD280 pKa = 3.42GTTTGRR286 pKa = 11.84ITSTGLHH293 pKa = 5.08TMKK296 pKa = 10.81KK297 pKa = 9.42LRR299 pKa = 11.84TMSWTTFPSSSVRR312 pKa = 11.84AGSSSSAVKK321 pKa = 9.1KK322 pKa = 6.46TTT324 pKa = 3.14
MM1 pKa = 6.97STSLSITSDD10 pKa = 2.59GRR12 pKa = 11.84HH13 pKa = 4.58SVRR16 pKa = 11.84SFRR19 pKa = 11.84HH20 pKa = 5.12RR21 pKa = 11.84NANTFLTYY29 pKa = 10.26SKK31 pKa = 10.88CPLEE35 pKa = 3.93PEE37 pKa = 4.86FIGEE41 pKa = 3.97HH42 pKa = 6.35LFRR45 pKa = 11.84LTKK48 pKa = 10.5DD49 pKa = 3.58FEE51 pKa = 4.03PAYY54 pKa = 9.19ILVVRR59 pKa = 11.84EE60 pKa = 3.81THH62 pKa = 6.89QDD64 pKa = 3.85GTWHH68 pKa = 6.13CHH70 pKa = 6.66ALLQCIKK77 pKa = 10.37PVTTRR82 pKa = 11.84DD83 pKa = 3.08EE84 pKa = 4.6RR85 pKa = 11.84YY86 pKa = 9.54FDD88 pKa = 3.44IDD90 pKa = 3.67RR91 pKa = 11.84YY92 pKa = 10.35HH93 pKa = 7.33PNIQSAKK100 pKa = 8.17STDD103 pKa = 3.09KK104 pKa = 10.55VRR106 pKa = 11.84EE107 pKa = 4.1YY108 pKa = 10.68ILKK111 pKa = 10.31DD112 pKa = 3.71PKK114 pKa = 11.09DD115 pKa = 3.29KK116 pKa = 10.5WEE118 pKa = 4.22KK119 pKa = 8.82GTYY122 pKa = 9.25IPRR125 pKa = 11.84KK126 pKa = 9.51KK127 pKa = 10.78SFVPPGKK134 pKa = 10.11EE135 pKa = 3.6NSEE138 pKa = 4.16KK139 pKa = 10.64KK140 pKa = 9.44PSKK143 pKa = 10.63DD144 pKa = 3.13EE145 pKa = 3.95VMKK148 pKa = 10.81EE149 pKa = 3.71IMTHH153 pKa = 4.37ATSRR157 pKa = 11.84AEE159 pKa = 3.89YY160 pKa = 10.09LSLVQTSLPYY170 pKa = 10.68DD171 pKa = 3.35WATKK175 pKa = 10.42LSYY178 pKa = 10.7FEE180 pKa = 4.77YY181 pKa = 10.06SASRR185 pKa = 11.84LFPDD189 pKa = 3.14IAEE192 pKa = 4.97PYY194 pKa = 9.71SNPHH198 pKa = 6.94PATDD202 pKa = 4.42PDD204 pKa = 4.21LLCNEE209 pKa = 4.77TLQDD213 pKa = 3.25WLEE216 pKa = 3.89PNIYY220 pKa = 9.6QVSPKK225 pKa = 10.59AYY227 pKa = 8.25MLLEE231 pKa = 4.21PSCLSLEE238 pKa = 4.12QAKK241 pKa = 10.7ADD243 pKa = 4.11LEE245 pKa = 4.43WLSEE249 pKa = 4.2TTRR252 pKa = 11.84LFQEE256 pKa = 4.37QEE258 pKa = 4.35SEE260 pKa = 4.12ASTSSAQHH268 pKa = 5.76GQVKK272 pKa = 10.26HH273 pKa = 6.34PGPEE277 pKa = 4.01ASDD280 pKa = 3.42GTTTGRR286 pKa = 11.84ITSTGLHH293 pKa = 5.08TMKK296 pKa = 10.81KK297 pKa = 9.42LRR299 pKa = 11.84TMSWTTFPSSSVRR312 pKa = 11.84AGSSSSAVKK321 pKa = 9.1KK322 pKa = 6.46TTT324 pKa = 3.14
Molecular weight: 36.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84370|Q84370_9GEMI Replication-associated protein OS=Panicum streak virus - Karino OX=268779 GN=RepA PE=3 SV=1
MM1 pKa = 7.63SGALKK6 pKa = 10.35RR7 pKa = 11.84KK8 pKa = 10.08RR9 pKa = 11.84SDD11 pKa = 2.87EE12 pKa = 4.38VAWSRR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.95PVKK22 pKa = 10.44KK23 pKa = 9.94QDD25 pKa = 3.19TGFPLPRR32 pKa = 11.84AGPSVRR38 pKa = 11.84RR39 pKa = 11.84GLPALQIQTLTAAGDD54 pKa = 3.66TMITVPSGGICSLIGTYY71 pKa = 10.73ARR73 pKa = 11.84GSDD76 pKa = 3.18EE77 pKa = 4.43GNRR80 pKa = 11.84HH81 pKa = 4.63TNEE84 pKa = 3.56TLTYY88 pKa = 10.08KK89 pKa = 10.59VALDD93 pKa = 3.36YY94 pKa = 11.55HH95 pKa = 6.49FVATAAACKK104 pKa = 9.83YY105 pKa = 9.96SSIGVGVMWLVYY117 pKa = 9.96DD118 pKa = 4.42AQPTGNSPEE127 pKa = 4.06VKK129 pKa = 10.2DD130 pKa = 4.13IFPHH134 pKa = 6.71SDD136 pKa = 2.91TLSAFPYY143 pKa = 7.04TWKK146 pKa = 10.5VGRR149 pKa = 11.84EE150 pKa = 3.83VCHH153 pKa = 6.21RR154 pKa = 11.84FVVKK158 pKa = 10.44RR159 pKa = 11.84RR160 pKa = 11.84WCFTMEE166 pKa = 3.97TNGRR170 pKa = 11.84IGSDD174 pKa = 3.35VPPANTAWPPCKK186 pKa = 9.69KK187 pKa = 10.32DD188 pKa = 3.04IYY190 pKa = 10.04FHH192 pKa = 6.77KK193 pKa = 10.19FCTGLGVKK201 pKa = 7.99TEE203 pKa = 4.15WKK205 pKa = 10.26NVTDD209 pKa = 4.33GKK211 pKa = 11.04VGAIKK216 pKa = 10.37KK217 pKa = 8.43GALYY221 pKa = 10.16IVIAPGNGLEE231 pKa = 4.27FTVHH235 pKa = 5.63GQCRR239 pKa = 11.84LYY241 pKa = 10.55FKK243 pKa = 10.94SVGNQQ248 pKa = 2.8
MM1 pKa = 7.63SGALKK6 pKa = 10.35RR7 pKa = 11.84KK8 pKa = 10.08RR9 pKa = 11.84SDD11 pKa = 2.87EE12 pKa = 4.38VAWSRR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.95PVKK22 pKa = 10.44KK23 pKa = 9.94QDD25 pKa = 3.19TGFPLPRR32 pKa = 11.84AGPSVRR38 pKa = 11.84RR39 pKa = 11.84GLPALQIQTLTAAGDD54 pKa = 3.66TMITVPSGGICSLIGTYY71 pKa = 10.73ARR73 pKa = 11.84GSDD76 pKa = 3.18EE77 pKa = 4.43GNRR80 pKa = 11.84HH81 pKa = 4.63TNEE84 pKa = 3.56TLTYY88 pKa = 10.08KK89 pKa = 10.59VALDD93 pKa = 3.36YY94 pKa = 11.55HH95 pKa = 6.49FVATAAACKK104 pKa = 9.83YY105 pKa = 9.96SSIGVGVMWLVYY117 pKa = 9.96DD118 pKa = 4.42AQPTGNSPEE127 pKa = 4.06VKK129 pKa = 10.2DD130 pKa = 4.13IFPHH134 pKa = 6.71SDD136 pKa = 2.91TLSAFPYY143 pKa = 7.04TWKK146 pKa = 10.5VGRR149 pKa = 11.84EE150 pKa = 3.83VCHH153 pKa = 6.21RR154 pKa = 11.84FVVKK158 pKa = 10.44RR159 pKa = 11.84RR160 pKa = 11.84WCFTMEE166 pKa = 3.97TNGRR170 pKa = 11.84IGSDD174 pKa = 3.35VPPANTAWPPCKK186 pKa = 9.69KK187 pKa = 10.32DD188 pKa = 3.04IYY190 pKa = 10.04FHH192 pKa = 6.77KK193 pKa = 10.19FCTGLGVKK201 pKa = 7.99TEE203 pKa = 4.15WKK205 pKa = 10.26NVTDD209 pKa = 4.33GKK211 pKa = 11.04VGAIKK216 pKa = 10.37KK217 pKa = 8.43GALYY221 pKa = 10.16IVIAPGNGLEE231 pKa = 4.27FTVHH235 pKa = 5.63GQCRR239 pKa = 11.84LYY241 pKa = 10.55FKK243 pKa = 10.94SVGNQQ248 pKa = 2.8
Molecular weight: 27.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
679 |
107 |
324 |
226.3 |
25.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.775 ± 0.817 | 2.062 ± 0.382 |
4.713 ± 0.385 | 5.449 ± 1.471 |
3.976 ± 0.517 | 6.48 ± 2.046 |
2.651 ± 0.876 | 3.976 ± 0.223 |
6.627 ± 1.345 | 7.658 ± 1.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.62 ± 0.213 | 2.209 ± 0.676 |
7.953 ± 2.242 | 2.798 ± 0.411 |
5.744 ± 0.345 | 9.131 ± 1.729 |
8.1 ± 1.356 | 6.48 ± 1.866 |
2.209 ± 0.25 | 3.387 ± 0.716 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
