
Salvia splendens (Scarlet sage)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salviinae; Salvia;
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
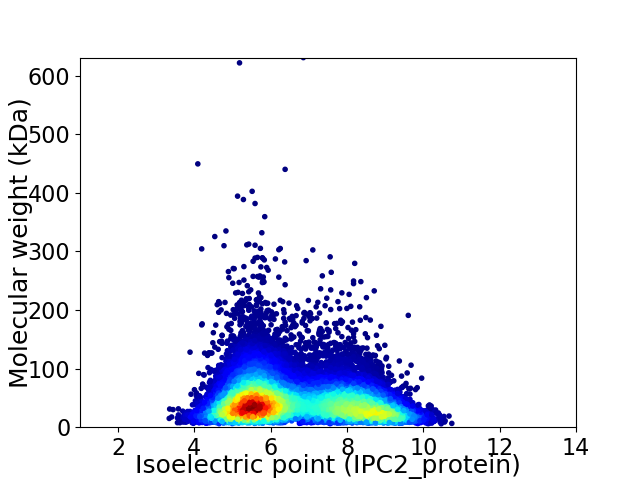
Virtual 2D-PAGE plot for 52672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D9A735|A0A4D9A735_SALSN Uncharacterized protein OS=Salvia splendens OX=180675 GN=Saspl_037418 PE=4 SV=1
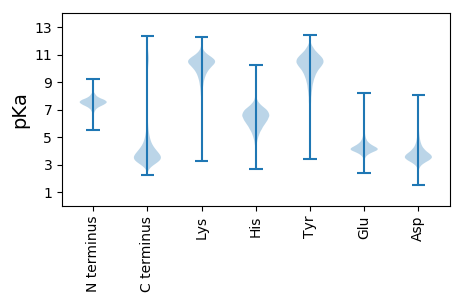
MM1 pKa = 7.76ALNNNLNLLFILLFFSSLLLPNHH24 pKa = 7.11LAQSPRR30 pKa = 11.84PAPGSSQGPAPGPSLYY46 pKa = 10.56DD47 pKa = 4.04DD48 pKa = 5.55EE49 pKa = 4.76PAPWPSLFDD58 pKa = 4.69DD59 pKa = 5.02EE60 pKa = 4.87PAPGPSLFDD69 pKa = 5.51DD70 pKa = 3.84IDD72 pKa = 3.97YY73 pKa = 11.27QIDD76 pKa = 3.73TQNPSISLPPTSSEE90 pKa = 4.23APSIAPSNEE99 pKa = 3.45YY100 pKa = 10.66DD101 pKa = 4.01LSPSNAPSDD110 pKa = 5.06DD111 pKa = 4.26DD112 pKa = 4.25PADD115 pKa = 3.78PALEE119 pKa = 4.6KK120 pKa = 10.38ICEE123 pKa = 4.48STDD126 pKa = 3.29HH127 pKa = 7.15PALCLSTVAPYY138 pKa = 11.03LDD140 pKa = 4.61GEE142 pKa = 4.39TDD144 pKa = 3.17IQSVLDD150 pKa = 3.57VAIQAGAVFSRR161 pKa = 11.84YY162 pKa = 8.64GQEE165 pKa = 4.12TAQKK169 pKa = 10.88LAMNPGNPPQHH180 pKa = 6.65SSVLSDD186 pKa = 3.51CRR188 pKa = 11.84DD189 pKa = 3.52GFEE192 pKa = 4.36TAAEE196 pKa = 4.32NYY198 pKa = 9.6GKK200 pKa = 10.46AADD203 pKa = 4.21ALAAQDD209 pKa = 3.6KK210 pKa = 9.44STVNSMLSAVITYY223 pKa = 10.12IGDD226 pKa = 3.83CQDD229 pKa = 4.08TISTDD234 pKa = 3.51SPLHH238 pKa = 5.93SLTDD242 pKa = 3.41KK243 pKa = 10.82LINMTSNCLAISSIINN259 pKa = 3.24
MM1 pKa = 7.76ALNNNLNLLFILLFFSSLLLPNHH24 pKa = 7.11LAQSPRR30 pKa = 11.84PAPGSSQGPAPGPSLYY46 pKa = 10.56DD47 pKa = 4.04DD48 pKa = 5.55EE49 pKa = 4.76PAPWPSLFDD58 pKa = 4.69DD59 pKa = 5.02EE60 pKa = 4.87PAPGPSLFDD69 pKa = 5.51DD70 pKa = 3.84IDD72 pKa = 3.97YY73 pKa = 11.27QIDD76 pKa = 3.73TQNPSISLPPTSSEE90 pKa = 4.23APSIAPSNEE99 pKa = 3.45YY100 pKa = 10.66DD101 pKa = 4.01LSPSNAPSDD110 pKa = 5.06DD111 pKa = 4.26DD112 pKa = 4.25PADD115 pKa = 3.78PALEE119 pKa = 4.6KK120 pKa = 10.38ICEE123 pKa = 4.48STDD126 pKa = 3.29HH127 pKa = 7.15PALCLSTVAPYY138 pKa = 11.03LDD140 pKa = 4.61GEE142 pKa = 4.39TDD144 pKa = 3.17IQSVLDD150 pKa = 3.57VAIQAGAVFSRR161 pKa = 11.84YY162 pKa = 8.64GQEE165 pKa = 4.12TAQKK169 pKa = 10.88LAMNPGNPPQHH180 pKa = 6.65SSVLSDD186 pKa = 3.51CRR188 pKa = 11.84DD189 pKa = 3.52GFEE192 pKa = 4.36TAAEE196 pKa = 4.32NYY198 pKa = 9.6GKK200 pKa = 10.46AADD203 pKa = 4.21ALAAQDD209 pKa = 3.6KK210 pKa = 9.44STVNSMLSAVITYY223 pKa = 10.12IGDD226 pKa = 3.83CQDD229 pKa = 4.08TISTDD234 pKa = 3.51SPLHH238 pKa = 5.93SLTDD242 pKa = 3.41KK243 pKa = 10.82LINMTSNCLAISSIINN259 pKa = 3.24
Molecular weight: 27.38 kDa
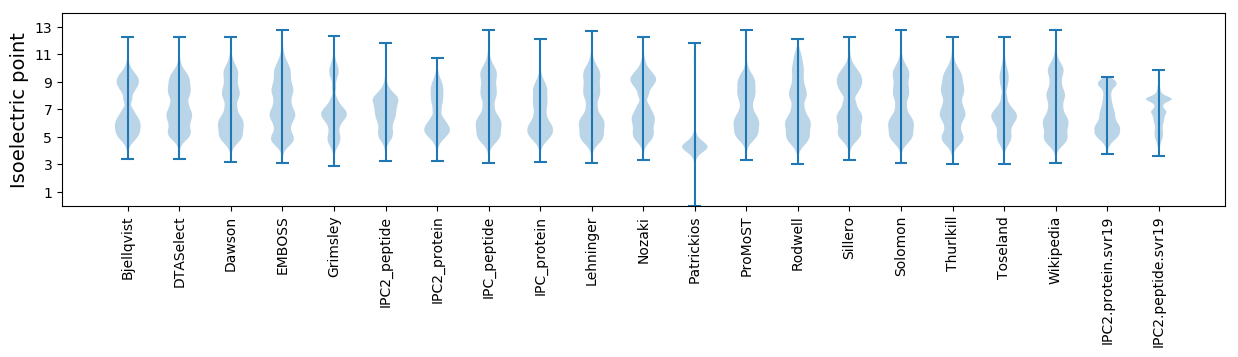
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D9AFZ0|A0A4D9AFZ0_SALSN Protein kinase domain-containing protein OS=Salvia splendens OX=180675 GN=Saspl_019786 PE=4 SV=1
MM1 pKa = 7.67RR2 pKa = 11.84CKK4 pKa = 10.3KK5 pKa = 9.86HH6 pKa = 6.67AADD9 pKa = 4.03LSSGVGVCASCLRR22 pKa = 11.84DD23 pKa = 3.22RR24 pKa = 11.84LFAIMEE30 pKa = 4.21AQALKK35 pKa = 10.46QSQQQQQQQNPPPLSFPRR53 pKa = 11.84SVSPYY58 pKa = 8.41ISRR61 pKa = 11.84RR62 pKa = 11.84PTDD65 pKa = 3.04TAAATWQIHH74 pKa = 4.82GYY76 pKa = 9.01EE77 pKa = 4.08RR78 pKa = 11.84SLYY81 pKa = 7.26TTPQLAPAAAGEE93 pKa = 4.04ISHH96 pKa = 6.61GKK98 pKa = 7.02GHH100 pKa = 6.76RR101 pKa = 11.84GGRR104 pKa = 11.84FSSLFSGLFRR114 pKa = 11.84SKK116 pKa = 10.84SDD118 pKa = 3.02KK119 pKa = 10.86HH120 pKa = 6.56DD121 pKa = 4.19FNSSSDD127 pKa = 3.44QGVSGQSCSASPFRR141 pKa = 11.84FSSMIPGRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84KK152 pKa = 9.6QIRR155 pKa = 11.84SFSVDD160 pKa = 2.99EE161 pKa = 4.27SSIGAQQSSCRR172 pKa = 11.84NRR174 pKa = 11.84DD175 pKa = 3.04RR176 pKa = 11.84GMSPARR182 pKa = 11.84CSDD185 pKa = 3.5EE186 pKa = 5.36EE187 pKa = 4.21EE188 pKa = 5.47DD189 pKa = 3.91EE190 pKa = 4.52HH191 pKa = 8.43CHH193 pKa = 6.08GGSSGYY199 pKa = 10.36SSEE202 pKa = 4.99SSQGWRR208 pKa = 11.84QTPRR212 pKa = 11.84RR213 pKa = 11.84TPARR217 pKa = 11.84SRR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84GGGGCQNRR229 pKa = 11.84NLSGLAFCLSPLVRR243 pKa = 11.84ASPSRR248 pKa = 11.84QWNQKK253 pKa = 9.37GIPPEE258 pKa = 3.88AVLPGEE264 pKa = 4.31TRR266 pKa = 11.84VPAKK270 pKa = 9.99PHH272 pKa = 6.02LSAAASFCKK281 pKa = 10.16NRR283 pKa = 11.84SRR285 pKa = 11.84KK286 pKa = 9.38LADD289 pKa = 3.75FGRR292 pKa = 11.84YY293 pKa = 7.44NSRR296 pKa = 11.84HH297 pKa = 4.49
MM1 pKa = 7.67RR2 pKa = 11.84CKK4 pKa = 10.3KK5 pKa = 9.86HH6 pKa = 6.67AADD9 pKa = 4.03LSSGVGVCASCLRR22 pKa = 11.84DD23 pKa = 3.22RR24 pKa = 11.84LFAIMEE30 pKa = 4.21AQALKK35 pKa = 10.46QSQQQQQQQNPPPLSFPRR53 pKa = 11.84SVSPYY58 pKa = 8.41ISRR61 pKa = 11.84RR62 pKa = 11.84PTDD65 pKa = 3.04TAAATWQIHH74 pKa = 4.82GYY76 pKa = 9.01EE77 pKa = 4.08RR78 pKa = 11.84SLYY81 pKa = 7.26TTPQLAPAAAGEE93 pKa = 4.04ISHH96 pKa = 6.61GKK98 pKa = 7.02GHH100 pKa = 6.76RR101 pKa = 11.84GGRR104 pKa = 11.84FSSLFSGLFRR114 pKa = 11.84SKK116 pKa = 10.84SDD118 pKa = 3.02KK119 pKa = 10.86HH120 pKa = 6.56DD121 pKa = 4.19FNSSSDD127 pKa = 3.44QGVSGQSCSASPFRR141 pKa = 11.84FSSMIPGRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84KK152 pKa = 9.6QIRR155 pKa = 11.84SFSVDD160 pKa = 2.99EE161 pKa = 4.27SSIGAQQSSCRR172 pKa = 11.84NRR174 pKa = 11.84DD175 pKa = 3.04RR176 pKa = 11.84GMSPARR182 pKa = 11.84CSDD185 pKa = 3.5EE186 pKa = 5.36EE187 pKa = 4.21EE188 pKa = 5.47DD189 pKa = 3.91EE190 pKa = 4.52HH191 pKa = 8.43CHH193 pKa = 6.08GGSSGYY199 pKa = 10.36SSEE202 pKa = 4.99SSQGWRR208 pKa = 11.84QTPRR212 pKa = 11.84RR213 pKa = 11.84TPARR217 pKa = 11.84SRR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84GGGGCQNRR229 pKa = 11.84NLSGLAFCLSPLVRR243 pKa = 11.84ASPSRR248 pKa = 11.84QWNQKK253 pKa = 9.37GIPPEE258 pKa = 3.88AVLPGEE264 pKa = 4.31TRR266 pKa = 11.84VPAKK270 pKa = 9.99PHH272 pKa = 6.02LSAAASFCKK281 pKa = 10.16NRR283 pKa = 11.84SRR285 pKa = 11.84KK286 pKa = 9.38LADD289 pKa = 3.75FGRR292 pKa = 11.84YY293 pKa = 7.44NSRR296 pKa = 11.84HH297 pKa = 4.49
Molecular weight: 32.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
22732906 |
50 |
5585 |
431.6 |
48.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.264 ± 0.01 | 1.859 ± 0.005 |
5.436 ± 0.007 | 6.534 ± 0.012 |
4.101 ± 0.007 | 6.613 ± 0.01 |
2.391 ± 0.005 | 5.173 ± 0.007 |
5.886 ± 0.01 | 9.692 ± 0.012 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.004 | 4.249 ± 0.007 |
4.91 ± 0.01 | 3.537 ± 0.007 |
5.523 ± 0.007 | 8.878 ± 0.013 |
4.742 ± 0.007 | 6.672 ± 0.008 |
1.27 ± 0.004 | 2.781 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
