
Capybara microvirus Cap1_SP_143
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.37
Get precalculated fractions of proteins
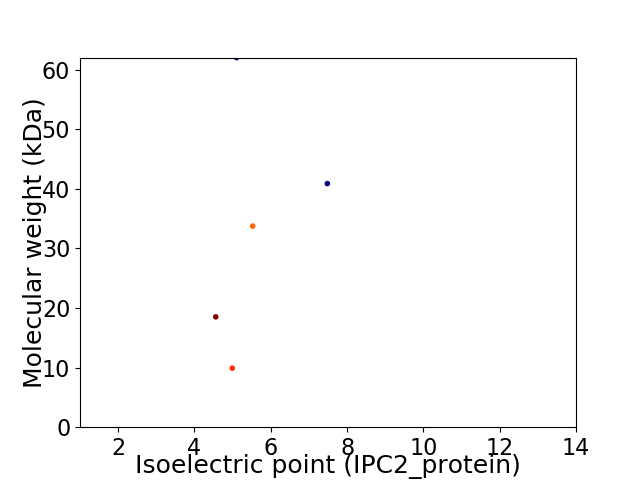
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
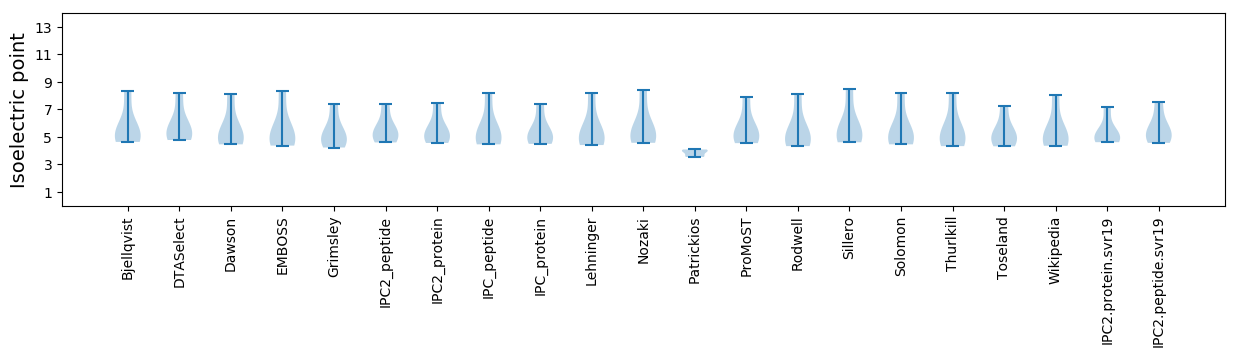
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVP9|A0A4V1FVP9_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_143 OX=2585389 PE=4 SV=1
MM1 pKa = 7.48LRR3 pKa = 11.84TNLTLSKK10 pKa = 10.45RR11 pKa = 11.84IFSDD15 pKa = 3.05SGSRR19 pKa = 11.84FVDD22 pKa = 4.01TYY24 pKa = 11.26RR25 pKa = 11.84PVINADD31 pKa = 2.61GHH33 pKa = 5.97MDD35 pKa = 5.32LIQSGKK41 pKa = 9.73KK42 pKa = 9.79DD43 pKa = 3.58VYY45 pKa = 11.61ADD47 pKa = 3.5IQSHH51 pKa = 6.66AEE53 pKa = 3.8EE54 pKa = 5.12CDD56 pKa = 3.18INLLIEE62 pKa = 4.86KK63 pKa = 9.85YY64 pKa = 10.94QMTGDD69 pKa = 3.49PAVINKK75 pKa = 9.78RR76 pKa = 11.84EE77 pKa = 3.97GLYY80 pKa = 10.69LDD82 pKa = 3.73VTDD85 pKa = 4.15MPKK88 pKa = 9.82TYY90 pKa = 10.94AEE92 pKa = 3.8IYY94 pKa = 10.02EE95 pKa = 4.37RR96 pKa = 11.84VAQAQDD102 pKa = 3.5VFSKK106 pKa = 11.01LPLSVKK112 pKa = 10.14EE113 pKa = 4.86KK114 pKa = 10.88FDD116 pKa = 4.02FDD118 pKa = 3.91PAKK121 pKa = 10.52FFASLGSKK129 pKa = 7.46EE130 pKa = 4.22TSALLAEE137 pKa = 4.76TFNASPASDD146 pKa = 4.08AGDD149 pKa = 3.67ASSSLSSNEE158 pKa = 3.51AATPTLSVEE167 pKa = 4.22GDD169 pKa = 3.15
MM1 pKa = 7.48LRR3 pKa = 11.84TNLTLSKK10 pKa = 10.45RR11 pKa = 11.84IFSDD15 pKa = 3.05SGSRR19 pKa = 11.84FVDD22 pKa = 4.01TYY24 pKa = 11.26RR25 pKa = 11.84PVINADD31 pKa = 2.61GHH33 pKa = 5.97MDD35 pKa = 5.32LIQSGKK41 pKa = 9.73KK42 pKa = 9.79DD43 pKa = 3.58VYY45 pKa = 11.61ADD47 pKa = 3.5IQSHH51 pKa = 6.66AEE53 pKa = 3.8EE54 pKa = 5.12CDD56 pKa = 3.18INLLIEE62 pKa = 4.86KK63 pKa = 9.85YY64 pKa = 10.94QMTGDD69 pKa = 3.49PAVINKK75 pKa = 9.78RR76 pKa = 11.84EE77 pKa = 3.97GLYY80 pKa = 10.69LDD82 pKa = 3.73VTDD85 pKa = 4.15MPKK88 pKa = 9.82TYY90 pKa = 10.94AEE92 pKa = 3.8IYY94 pKa = 10.02EE95 pKa = 4.37RR96 pKa = 11.84VAQAQDD102 pKa = 3.5VFSKK106 pKa = 11.01LPLSVKK112 pKa = 10.14EE113 pKa = 4.86KK114 pKa = 10.88FDD116 pKa = 4.02FDD118 pKa = 3.91PAKK121 pKa = 10.52FFASLGSKK129 pKa = 7.46EE130 pKa = 4.22TSALLAEE137 pKa = 4.76TFNASPASDD146 pKa = 4.08AGDD149 pKa = 3.67ASSSLSSNEE158 pKa = 3.51AATPTLSVEE167 pKa = 4.22GDD169 pKa = 3.15
Molecular weight: 18.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7Z4|A0A4P8W7Z4_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_143 OX=2585389 PE=3 SV=1
MM1 pKa = 7.52ACSNPNRR8 pKa = 11.84IFVIGKK14 pKa = 8.72NPKK17 pKa = 8.28TGKK20 pKa = 10.14KK21 pKa = 10.11KK22 pKa = 10.3IVFCSRR28 pKa = 11.84FVRR31 pKa = 11.84AVGVKK36 pKa = 10.36DD37 pKa = 3.94DD38 pKa = 4.04GSVDD42 pKa = 3.26RR43 pKa = 11.84FYY45 pKa = 11.46EE46 pKa = 4.34DD47 pKa = 3.56PEE49 pKa = 4.17KK50 pKa = 11.34DD51 pKa = 3.37KK52 pKa = 11.76VPFTYY57 pKa = 10.09RR58 pKa = 11.84SRR60 pKa = 11.84LYY62 pKa = 10.56KK63 pKa = 10.35FVVSDD68 pKa = 4.62YY69 pKa = 11.25IDD71 pKa = 3.87VPCGKK76 pKa = 10.27CLGCRR81 pKa = 11.84LAYY84 pKa = 10.14SKK86 pKa = 10.63QWTSRR91 pKa = 11.84IMLEE95 pKa = 4.74LPYY98 pKa = 9.92HH99 pKa = 5.37DD100 pKa = 5.01HH101 pKa = 6.18SWFVTLTYY109 pKa = 10.78DD110 pKa = 5.53DD111 pKa = 3.66EE112 pKa = 4.6HH113 pKa = 8.71LPINEE118 pKa = 3.95FTDD121 pKa = 3.93PVTSEE126 pKa = 4.12VGEE129 pKa = 4.24STTLVKK135 pKa = 10.52RR136 pKa = 11.84HH137 pKa = 5.37VQLFMKK143 pKa = 10.1KK144 pKa = 9.78LRR146 pKa = 11.84NAYY149 pKa = 8.88PDD151 pKa = 3.15NHH153 pKa = 7.27LMYY156 pKa = 10.2FFAGEE161 pKa = 3.89YY162 pKa = 9.88GDD164 pKa = 3.57RR165 pKa = 11.84SRR167 pKa = 11.84RR168 pKa = 11.84PHH170 pKa = 4.59YY171 pKa = 10.08HH172 pKa = 7.17AIIFDD177 pKa = 4.31LPLDD181 pKa = 4.04DD182 pKa = 4.97LVFYY186 pKa = 8.76KK187 pKa = 10.47TSPNGDD193 pKa = 2.98IYY195 pKa = 11.22FNSPKK200 pKa = 10.35LEE202 pKa = 4.65KK203 pKa = 9.62IWSHH207 pKa = 5.58GFVVVANVTYY217 pKa = 10.99ASAAYY222 pKa = 5.99TARR225 pKa = 11.84YY226 pKa = 8.85IMKK229 pKa = 9.18KK230 pKa = 10.34QNVSDD235 pKa = 3.96EE236 pKa = 3.91NSIYY240 pKa = 10.52ARR242 pKa = 11.84KK243 pKa = 10.05NIEE246 pKa = 3.92PEE248 pKa = 3.99FCLMSQKK255 pKa = 10.41PSIGRR260 pKa = 11.84RR261 pKa = 11.84YY262 pKa = 10.21FEE264 pKa = 4.82DD265 pKa = 3.92HH266 pKa = 7.27PDD268 pKa = 3.77VYY270 pKa = 11.44DD271 pKa = 4.65LDD273 pKa = 4.24YY274 pKa = 11.5LSLAGGYY281 pKa = 8.48RR282 pKa = 11.84ASIPKK287 pKa = 10.04YY288 pKa = 10.14FDD290 pKa = 3.42YY291 pKa = 11.28LLEE294 pKa = 4.44KK295 pKa = 10.55VDD297 pKa = 4.35AEE299 pKa = 3.82RR300 pKa = 11.84LQAIKK305 pKa = 10.36EE306 pKa = 4.01EE307 pKa = 4.24RR308 pKa = 11.84KK309 pKa = 9.94FFAQHH314 pKa = 6.4LANIKK319 pKa = 10.36EE320 pKa = 4.3SMHH323 pKa = 5.7TYY325 pKa = 10.4DD326 pKa = 4.71RR327 pKa = 11.84EE328 pKa = 4.23SHH330 pKa = 6.52FDD332 pKa = 3.69VIHH335 pKa = 6.61EE336 pKa = 4.21NLVSRR341 pKa = 11.84SKK343 pKa = 10.94HH344 pKa = 4.79LAKK347 pKa = 10.69DD348 pKa = 3.56LL349 pKa = 4.02
MM1 pKa = 7.52ACSNPNRR8 pKa = 11.84IFVIGKK14 pKa = 8.72NPKK17 pKa = 8.28TGKK20 pKa = 10.14KK21 pKa = 10.11KK22 pKa = 10.3IVFCSRR28 pKa = 11.84FVRR31 pKa = 11.84AVGVKK36 pKa = 10.36DD37 pKa = 3.94DD38 pKa = 4.04GSVDD42 pKa = 3.26RR43 pKa = 11.84FYY45 pKa = 11.46EE46 pKa = 4.34DD47 pKa = 3.56PEE49 pKa = 4.17KK50 pKa = 11.34DD51 pKa = 3.37KK52 pKa = 11.76VPFTYY57 pKa = 10.09RR58 pKa = 11.84SRR60 pKa = 11.84LYY62 pKa = 10.56KK63 pKa = 10.35FVVSDD68 pKa = 4.62YY69 pKa = 11.25IDD71 pKa = 3.87VPCGKK76 pKa = 10.27CLGCRR81 pKa = 11.84LAYY84 pKa = 10.14SKK86 pKa = 10.63QWTSRR91 pKa = 11.84IMLEE95 pKa = 4.74LPYY98 pKa = 9.92HH99 pKa = 5.37DD100 pKa = 5.01HH101 pKa = 6.18SWFVTLTYY109 pKa = 10.78DD110 pKa = 5.53DD111 pKa = 3.66EE112 pKa = 4.6HH113 pKa = 8.71LPINEE118 pKa = 3.95FTDD121 pKa = 3.93PVTSEE126 pKa = 4.12VGEE129 pKa = 4.24STTLVKK135 pKa = 10.52RR136 pKa = 11.84HH137 pKa = 5.37VQLFMKK143 pKa = 10.1KK144 pKa = 9.78LRR146 pKa = 11.84NAYY149 pKa = 8.88PDD151 pKa = 3.15NHH153 pKa = 7.27LMYY156 pKa = 10.2FFAGEE161 pKa = 3.89YY162 pKa = 9.88GDD164 pKa = 3.57RR165 pKa = 11.84SRR167 pKa = 11.84RR168 pKa = 11.84PHH170 pKa = 4.59YY171 pKa = 10.08HH172 pKa = 7.17AIIFDD177 pKa = 4.31LPLDD181 pKa = 4.04DD182 pKa = 4.97LVFYY186 pKa = 8.76KK187 pKa = 10.47TSPNGDD193 pKa = 2.98IYY195 pKa = 11.22FNSPKK200 pKa = 10.35LEE202 pKa = 4.65KK203 pKa = 9.62IWSHH207 pKa = 5.58GFVVVANVTYY217 pKa = 10.99ASAAYY222 pKa = 5.99TARR225 pKa = 11.84YY226 pKa = 8.85IMKK229 pKa = 9.18KK230 pKa = 10.34QNVSDD235 pKa = 3.96EE236 pKa = 3.91NSIYY240 pKa = 10.52ARR242 pKa = 11.84KK243 pKa = 10.05NIEE246 pKa = 3.92PEE248 pKa = 3.99FCLMSQKK255 pKa = 10.41PSIGRR260 pKa = 11.84RR261 pKa = 11.84YY262 pKa = 10.21FEE264 pKa = 4.82DD265 pKa = 3.92HH266 pKa = 7.27PDD268 pKa = 3.77VYY270 pKa = 11.44DD271 pKa = 4.65LDD273 pKa = 4.24YY274 pKa = 11.5LSLAGGYY281 pKa = 8.48RR282 pKa = 11.84ASIPKK287 pKa = 10.04YY288 pKa = 10.14FDD290 pKa = 3.42YY291 pKa = 11.28LLEE294 pKa = 4.44KK295 pKa = 10.55VDD297 pKa = 4.35AEE299 pKa = 3.82RR300 pKa = 11.84LQAIKK305 pKa = 10.36EE306 pKa = 4.01EE307 pKa = 4.24RR308 pKa = 11.84KK309 pKa = 9.94FFAQHH314 pKa = 6.4LANIKK319 pKa = 10.36EE320 pKa = 4.3SMHH323 pKa = 5.7TYY325 pKa = 10.4DD326 pKa = 4.71RR327 pKa = 11.84EE328 pKa = 4.23SHH330 pKa = 6.52FDD332 pKa = 3.69VIHH335 pKa = 6.61EE336 pKa = 4.21NLVSRR341 pKa = 11.84SKK343 pKa = 10.94HH344 pKa = 4.79LAKK347 pKa = 10.69DD348 pKa = 3.56LL349 pKa = 4.02
Molecular weight: 40.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1478 |
87 |
551 |
295.6 |
33.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.593 ± 2.225 | 0.609 ± 0.338 |
6.428 ± 0.975 | 4.736 ± 0.578 |
4.736 ± 0.98 | 5.683 ± 0.776 |
2.165 ± 0.67 | 5.074 ± 0.704 |
4.804 ± 1.016 | 7.172 ± 0.617 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.724 | 6.157 ± 1.564 |
4.668 ± 0.612 | 3.518 ± 0.552 |
4.398 ± 0.696 | 10.352 ± 1.6 |
5.548 ± 0.594 | 6.089 ± 0.579 |
1.218 ± 0.397 | 5.413 ± 0.582 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
