
Primula malacoides virus China/Mar2007
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus; Primula malacoides virus 1
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
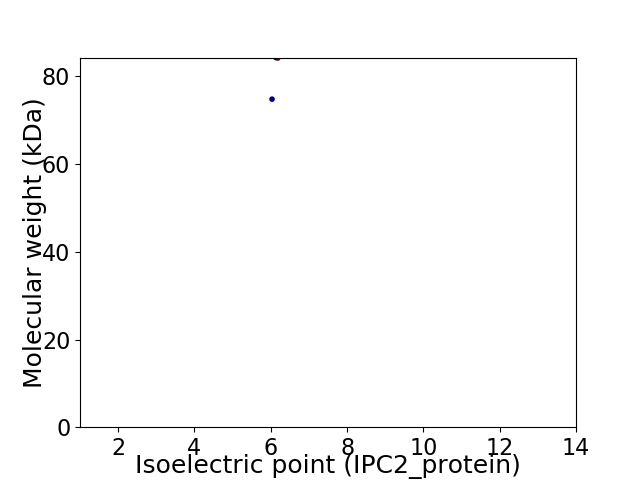
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8W7G2|A8W7G2_9VIRU Putative capsid protein OS=Primula malacoides virus China/Mar2007 OX=479713 PE=4 SV=1
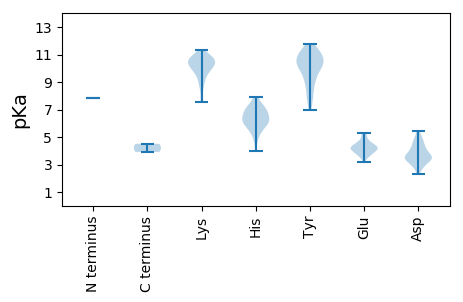
MM1 pKa = 7.81PFNAVRR7 pKa = 11.84NYY9 pKa = 10.0LAEE12 pKa = 3.97RR13 pKa = 11.84LVRR16 pKa = 11.84VRR18 pKa = 11.84TEE20 pKa = 3.93LKK22 pKa = 10.74NFTSSNRR29 pKa = 11.84EE30 pKa = 3.18PDD32 pKa = 3.15ATLEE36 pKa = 4.05LSQDD40 pKa = 3.3PDD42 pKa = 2.96LRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 9.82YY47 pKa = 11.24DD48 pKa = 3.19NTRR51 pKa = 11.84YY52 pKa = 10.27NSSNDD57 pKa = 2.93AKK59 pKa = 10.98YY60 pKa = 8.34RR61 pKa = 11.84TLNKK65 pKa = 10.05EE66 pKa = 3.51YY67 pKa = 9.43STLVEE72 pKa = 5.25AYY74 pKa = 8.27RR75 pKa = 11.84TDD77 pKa = 3.38NQQKK81 pKa = 7.73HH82 pKa = 4.96QPYY85 pKa = 9.34EE86 pKa = 3.75LHH88 pKa = 6.15QPIPADD94 pKa = 3.47AAPIIDD100 pKa = 4.23KK101 pKa = 10.31RR102 pKa = 11.84QPAPGLKK109 pKa = 9.6LVPLMYY115 pKa = 10.21HH116 pKa = 6.08YY117 pKa = 11.23GHH119 pKa = 7.13VIHH122 pKa = 7.49DD123 pKa = 4.88PDD125 pKa = 5.4PDD127 pKa = 3.61QPDD130 pKa = 3.36QTKK133 pKa = 10.21VYY135 pKa = 9.33PLDD138 pKa = 3.46SRR140 pKa = 11.84IYY142 pKa = 8.53NLIMSTYY149 pKa = 8.84PSYY152 pKa = 11.38LSVLHH157 pKa = 6.91DD158 pKa = 3.61YY159 pKa = 10.18CRR161 pKa = 11.84PIGTVEE167 pKa = 3.7ATFNDD172 pKa = 4.12FNTEE176 pKa = 4.13QIPSAPIDD184 pKa = 3.79EE185 pKa = 4.82DD186 pKa = 3.66RR187 pKa = 11.84KK188 pKa = 9.64QQILKK193 pKa = 10.22HH194 pKa = 5.58LFKK197 pKa = 10.95FLDD200 pKa = 3.63VKK202 pKa = 10.69PYY204 pKa = 10.98LPIHH208 pKa = 6.15FVDD211 pKa = 3.64TQYY214 pKa = 11.57CKK216 pKa = 10.19TPLVTGTGYY225 pKa = 10.81HH226 pKa = 5.59NRR228 pKa = 11.84YY229 pKa = 9.38SFKK232 pKa = 10.55QRR234 pKa = 11.84AHH236 pKa = 6.38AKK238 pKa = 8.37YY239 pKa = 10.09SHH241 pKa = 6.76PEE243 pKa = 3.6EE244 pKa = 4.37YY245 pKa = 10.6ALKK248 pKa = 8.8STSKK252 pKa = 10.77GYY254 pKa = 10.32FYY256 pKa = 11.17NATYY260 pKa = 10.79EE261 pKa = 3.99NARR264 pKa = 11.84TLVHH268 pKa = 7.17LIKK271 pKa = 10.08TYY273 pKa = 10.8GLPFNMQFACPKK285 pKa = 10.43SDD287 pKa = 4.26LTDD290 pKa = 3.52EE291 pKa = 4.48QINLYY296 pKa = 9.73ISKK299 pKa = 10.67ANQFFNDD306 pKa = 3.6YY307 pKa = 7.51PTLLFTRR314 pKa = 11.84NHH316 pKa = 5.35ISKK319 pKa = 10.63RR320 pKa = 11.84SGTLKK325 pKa = 10.07VRR327 pKa = 11.84PVYY330 pKa = 10.59AVDD333 pKa = 4.8DD334 pKa = 3.66IFIIIEE340 pKa = 4.07LMLTFPLTIQARR352 pKa = 11.84KK353 pKa = 8.74PSCCIMYY360 pKa = 10.42GLEE363 pKa = 4.17TIRR366 pKa = 11.84GSNHH370 pKa = 6.08YY371 pKa = 9.39IDD373 pKa = 4.88RR374 pKa = 11.84LARR377 pKa = 11.84PYY379 pKa = 10.35STFFSLDD386 pKa = 2.74WSSYY390 pKa = 6.99DD391 pKa = 3.17QRR393 pKa = 11.84LPRR396 pKa = 11.84VITDD400 pKa = 3.3IYY402 pKa = 9.28YY403 pKa = 9.62TDD405 pKa = 4.32FLRR408 pKa = 11.84SLIVINHH415 pKa = 7.09GYY417 pKa = 9.18QPTYY421 pKa = 10.39EE422 pKa = 4.22YY423 pKa = 8.37PTYY426 pKa = 10.44PDD428 pKa = 3.72LDD430 pKa = 3.56EE431 pKa = 5.2HH432 pKa = 6.91KK433 pKa = 10.43LYY435 pKa = 11.32SRR437 pKa = 11.84MDD439 pKa = 3.37NLLYY443 pKa = 10.37FLHH446 pKa = 6.14TWYY449 pKa = 11.35NNMTFLLPDD458 pKa = 4.16GYY460 pKa = 11.15AYY462 pKa = 10.68RR463 pKa = 11.84RR464 pKa = 11.84THH466 pKa = 6.4CGVPSGLYY474 pKa = 7.56NTQYY478 pKa = 11.31LDD480 pKa = 3.35SFGNLFLIIDD490 pKa = 3.86AMIEE494 pKa = 4.0FEE496 pKa = 4.25FTDD499 pKa = 3.95DD500 pKa = 4.72EE501 pKa = 4.35ISKK504 pKa = 9.98FQLLILGGDD513 pKa = 3.62NTGMTNLAIDD523 pKa = 5.24RR524 pKa = 11.84IDD526 pKa = 3.36KK527 pKa = 10.75FITFLEE533 pKa = 5.07TYY535 pKa = 10.78ALARR539 pKa = 11.84YY540 pKa = 10.2NMVLPKK546 pKa = 9.26TKK548 pKa = 10.53CILTSLRR555 pKa = 11.84SKK557 pKa = 10.46IVTLGYY563 pKa = 9.02EE564 pKa = 4.3CNYY567 pKa = 10.2GSPKK571 pKa = 9.94RR572 pKa = 11.84DD573 pKa = 2.66IHH575 pKa = 7.87KK576 pKa = 10.53LVAQLCFPEE585 pKa = 5.29NGLKK589 pKa = 10.23AHH591 pKa = 5.92TMSARR596 pKa = 11.84ATGIAYY602 pKa = 9.9ASAGQDD608 pKa = 2.64IMFHH612 pKa = 5.81SFCQDD617 pKa = 2.32VYY619 pKa = 11.4NIFRR623 pKa = 11.84SDD625 pKa = 3.49YY626 pKa = 10.75KK627 pKa = 10.75PDD629 pKa = 3.1VRR631 pKa = 11.84ANLYY635 pKa = 7.78FQRR638 pKa = 11.84QFLNDD643 pKa = 3.81LEE645 pKa = 4.72EE646 pKa = 4.49GVPDD650 pKa = 4.51LATPTVPPFPSLYY663 pKa = 9.97EE664 pKa = 3.71IRR666 pKa = 11.84KK667 pKa = 9.74LYY669 pKa = 10.74SKK671 pKa = 10.8YY672 pKa = 10.09QGPLSFTPKK681 pKa = 9.09WNSAHH686 pKa = 7.3FINEE690 pKa = 3.97PDD692 pKa = 3.82SVPISAKK699 pKa = 7.56TMRR702 pKa = 11.84QFEE705 pKa = 4.57EE706 pKa = 4.47EE707 pKa = 3.96FSIPLMTAATFEE719 pKa = 4.64TVVPP723 pKa = 4.51
MM1 pKa = 7.81PFNAVRR7 pKa = 11.84NYY9 pKa = 10.0LAEE12 pKa = 3.97RR13 pKa = 11.84LVRR16 pKa = 11.84VRR18 pKa = 11.84TEE20 pKa = 3.93LKK22 pKa = 10.74NFTSSNRR29 pKa = 11.84EE30 pKa = 3.18PDD32 pKa = 3.15ATLEE36 pKa = 4.05LSQDD40 pKa = 3.3PDD42 pKa = 2.96LRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 9.82YY47 pKa = 11.24DD48 pKa = 3.19NTRR51 pKa = 11.84YY52 pKa = 10.27NSSNDD57 pKa = 2.93AKK59 pKa = 10.98YY60 pKa = 8.34RR61 pKa = 11.84TLNKK65 pKa = 10.05EE66 pKa = 3.51YY67 pKa = 9.43STLVEE72 pKa = 5.25AYY74 pKa = 8.27RR75 pKa = 11.84TDD77 pKa = 3.38NQQKK81 pKa = 7.73HH82 pKa = 4.96QPYY85 pKa = 9.34EE86 pKa = 3.75LHH88 pKa = 6.15QPIPADD94 pKa = 3.47AAPIIDD100 pKa = 4.23KK101 pKa = 10.31RR102 pKa = 11.84QPAPGLKK109 pKa = 9.6LVPLMYY115 pKa = 10.21HH116 pKa = 6.08YY117 pKa = 11.23GHH119 pKa = 7.13VIHH122 pKa = 7.49DD123 pKa = 4.88PDD125 pKa = 5.4PDD127 pKa = 3.61QPDD130 pKa = 3.36QTKK133 pKa = 10.21VYY135 pKa = 9.33PLDD138 pKa = 3.46SRR140 pKa = 11.84IYY142 pKa = 8.53NLIMSTYY149 pKa = 8.84PSYY152 pKa = 11.38LSVLHH157 pKa = 6.91DD158 pKa = 3.61YY159 pKa = 10.18CRR161 pKa = 11.84PIGTVEE167 pKa = 3.7ATFNDD172 pKa = 4.12FNTEE176 pKa = 4.13QIPSAPIDD184 pKa = 3.79EE185 pKa = 4.82DD186 pKa = 3.66RR187 pKa = 11.84KK188 pKa = 9.64QQILKK193 pKa = 10.22HH194 pKa = 5.58LFKK197 pKa = 10.95FLDD200 pKa = 3.63VKK202 pKa = 10.69PYY204 pKa = 10.98LPIHH208 pKa = 6.15FVDD211 pKa = 3.64TQYY214 pKa = 11.57CKK216 pKa = 10.19TPLVTGTGYY225 pKa = 10.81HH226 pKa = 5.59NRR228 pKa = 11.84YY229 pKa = 9.38SFKK232 pKa = 10.55QRR234 pKa = 11.84AHH236 pKa = 6.38AKK238 pKa = 8.37YY239 pKa = 10.09SHH241 pKa = 6.76PEE243 pKa = 3.6EE244 pKa = 4.37YY245 pKa = 10.6ALKK248 pKa = 8.8STSKK252 pKa = 10.77GYY254 pKa = 10.32FYY256 pKa = 11.17NATYY260 pKa = 10.79EE261 pKa = 3.99NARR264 pKa = 11.84TLVHH268 pKa = 7.17LIKK271 pKa = 10.08TYY273 pKa = 10.8GLPFNMQFACPKK285 pKa = 10.43SDD287 pKa = 4.26LTDD290 pKa = 3.52EE291 pKa = 4.48QINLYY296 pKa = 9.73ISKK299 pKa = 10.67ANQFFNDD306 pKa = 3.6YY307 pKa = 7.51PTLLFTRR314 pKa = 11.84NHH316 pKa = 5.35ISKK319 pKa = 10.63RR320 pKa = 11.84SGTLKK325 pKa = 10.07VRR327 pKa = 11.84PVYY330 pKa = 10.59AVDD333 pKa = 4.8DD334 pKa = 3.66IFIIIEE340 pKa = 4.07LMLTFPLTIQARR352 pKa = 11.84KK353 pKa = 8.74PSCCIMYY360 pKa = 10.42GLEE363 pKa = 4.17TIRR366 pKa = 11.84GSNHH370 pKa = 6.08YY371 pKa = 9.39IDD373 pKa = 4.88RR374 pKa = 11.84LARR377 pKa = 11.84PYY379 pKa = 10.35STFFSLDD386 pKa = 2.74WSSYY390 pKa = 6.99DD391 pKa = 3.17QRR393 pKa = 11.84LPRR396 pKa = 11.84VITDD400 pKa = 3.3IYY402 pKa = 9.28YY403 pKa = 9.62TDD405 pKa = 4.32FLRR408 pKa = 11.84SLIVINHH415 pKa = 7.09GYY417 pKa = 9.18QPTYY421 pKa = 10.39EE422 pKa = 4.22YY423 pKa = 8.37PTYY426 pKa = 10.44PDD428 pKa = 3.72LDD430 pKa = 3.56EE431 pKa = 5.2HH432 pKa = 6.91KK433 pKa = 10.43LYY435 pKa = 11.32SRR437 pKa = 11.84MDD439 pKa = 3.37NLLYY443 pKa = 10.37FLHH446 pKa = 6.14TWYY449 pKa = 11.35NNMTFLLPDD458 pKa = 4.16GYY460 pKa = 11.15AYY462 pKa = 10.68RR463 pKa = 11.84RR464 pKa = 11.84THH466 pKa = 6.4CGVPSGLYY474 pKa = 7.56NTQYY478 pKa = 11.31LDD480 pKa = 3.35SFGNLFLIIDD490 pKa = 3.86AMIEE494 pKa = 4.0FEE496 pKa = 4.25FTDD499 pKa = 3.95DD500 pKa = 4.72EE501 pKa = 4.35ISKK504 pKa = 9.98FQLLILGGDD513 pKa = 3.62NTGMTNLAIDD523 pKa = 5.24RR524 pKa = 11.84IDD526 pKa = 3.36KK527 pKa = 10.75FITFLEE533 pKa = 5.07TYY535 pKa = 10.78ALARR539 pKa = 11.84YY540 pKa = 10.2NMVLPKK546 pKa = 9.26TKK548 pKa = 10.53CILTSLRR555 pKa = 11.84SKK557 pKa = 10.46IVTLGYY563 pKa = 9.02EE564 pKa = 4.3CNYY567 pKa = 10.2GSPKK571 pKa = 9.94RR572 pKa = 11.84DD573 pKa = 2.66IHH575 pKa = 7.87KK576 pKa = 10.53LVAQLCFPEE585 pKa = 5.29NGLKK589 pKa = 10.23AHH591 pKa = 5.92TMSARR596 pKa = 11.84ATGIAYY602 pKa = 9.9ASAGQDD608 pKa = 2.64IMFHH612 pKa = 5.81SFCQDD617 pKa = 2.32VYY619 pKa = 11.4NIFRR623 pKa = 11.84SDD625 pKa = 3.49YY626 pKa = 10.75KK627 pKa = 10.75PDD629 pKa = 3.1VRR631 pKa = 11.84ANLYY635 pKa = 7.78FQRR638 pKa = 11.84QFLNDD643 pKa = 3.81LEE645 pKa = 4.72EE646 pKa = 4.49GVPDD650 pKa = 4.51LATPTVPPFPSLYY663 pKa = 9.97EE664 pKa = 3.71IRR666 pKa = 11.84KK667 pKa = 9.74LYY669 pKa = 10.74SKK671 pKa = 10.8YY672 pKa = 10.09QGPLSFTPKK681 pKa = 9.09WNSAHH686 pKa = 7.3FINEE690 pKa = 3.97PDD692 pKa = 3.82SVPISAKK699 pKa = 7.56TMRR702 pKa = 11.84QFEE705 pKa = 4.57EE706 pKa = 4.47EE707 pKa = 3.96FSIPLMTAATFEE719 pKa = 4.64TVVPP723 pKa = 4.51
Molecular weight: 84.19 kDa
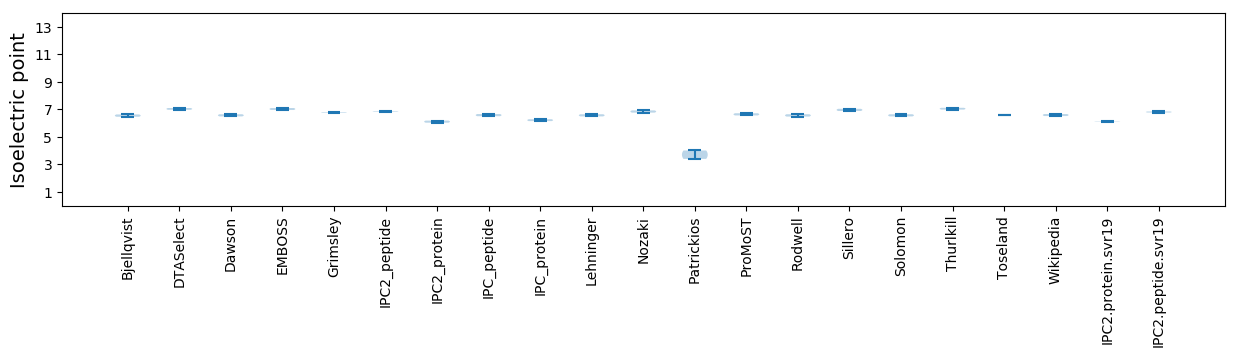
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8W7G2|A8W7G2_9VIRU Putative capsid protein OS=Primula malacoides virus China/Mar2007 OX=479713 PE=4 SV=1
MM1 pKa = 7.81SDD3 pKa = 2.97TVLQAKK9 pKa = 8.92TNFLKK14 pKa = 10.97NKK16 pKa = 10.01SPVLSVQEE24 pKa = 4.15SKK26 pKa = 10.7RR27 pKa = 11.84PKK29 pKa = 9.63ISRR32 pKa = 11.84DD33 pKa = 3.41LMPLFQSNTAARR45 pKa = 11.84SDD47 pKa = 4.47SSNQWTIDD55 pKa = 3.26VFPNMVPITMYY66 pKa = 11.06IMLTSIAHH74 pKa = 6.28ANDD77 pKa = 2.82VDD79 pKa = 3.51HH80 pKa = 6.89RR81 pKa = 11.84TTSKK85 pKa = 10.83VSVATICMYY94 pKa = 10.53HH95 pKa = 7.74LSIVYY100 pKa = 9.77GYY102 pKa = 10.95FLLNDD107 pKa = 4.23MITRR111 pKa = 11.84ASASAHH117 pKa = 4.79ARR119 pKa = 11.84PWIEE123 pKa = 3.89TSWKK127 pKa = 10.46RR128 pKa = 11.84EE129 pKa = 3.71FADD132 pKa = 4.66FLLTLPVPEE141 pKa = 4.83FLTPFLSQFQHH152 pKa = 5.65FTTDD156 pKa = 2.65RR157 pKa = 11.84TKK159 pKa = 11.2NVFLTPSAAGFDD171 pKa = 3.75HH172 pKa = 7.12NIFFGRR178 pKa = 11.84VFPLNMFSAIHH189 pKa = 7.12DD190 pKa = 4.03CTATLPGNTPRR201 pKa = 11.84IQVLQDD207 pKa = 3.45LYY209 pKa = 11.79SRR211 pKa = 11.84VLYY214 pKa = 9.92TITAPGFTCLIPDD227 pKa = 5.29LIGVTIDD234 pKa = 3.09QTTPTTANYY243 pKa = 7.91MNSKK247 pKa = 10.1LYY249 pKa = 10.06QVYY252 pKa = 7.91TAVFNPVLFRR262 pKa = 11.84DD263 pKa = 4.41FQHH266 pKa = 6.84RR267 pKa = 11.84SSLAALSFQSPVFANNHH284 pKa = 4.01VNAYY288 pKa = 10.02DD289 pKa = 3.97LLFSASAANLRR300 pKa = 11.84EE301 pKa = 4.29LKK303 pKa = 10.65VVLQVVSTVLDD314 pKa = 4.01GKK316 pKa = 9.33VTTSGTLGDD325 pKa = 4.83FLTKK329 pKa = 10.28PSGALAIKK337 pKa = 10.15HH338 pKa = 6.07GYY340 pKa = 7.32STYY343 pKa = 11.44ALPTWSYY350 pKa = 8.77NTNATKK356 pKa = 10.53SAVYY360 pKa = 10.16SAITTLTLTSEE371 pKa = 4.19RR372 pKa = 11.84DD373 pKa = 3.3RR374 pKa = 11.84AQDD377 pKa = 2.67ICFLQRR383 pKa = 11.84PAAAFNATTEE393 pKa = 4.39VTDD396 pKa = 3.3VTYY399 pKa = 10.79AATATPNQAVQLPTGHH415 pKa = 6.38QLTRR419 pKa = 11.84HH420 pKa = 5.74FPFSLLQVPNGAEE433 pKa = 3.69PFPRR437 pKa = 11.84HH438 pKa = 6.69DD439 pKa = 4.47NDD441 pKa = 5.27DD442 pKa = 3.81LVQFSDD448 pKa = 4.81FVHH451 pKa = 6.43TAPSVLVLDD460 pKa = 4.18TDD462 pKa = 4.33GEE464 pKa = 4.52LTISAHH470 pKa = 6.48LPLLAGKK477 pKa = 9.71IIEE480 pKa = 4.27SFEE483 pKa = 4.19IDD485 pKa = 3.09GSTIEE490 pKa = 4.59MPNVDD495 pKa = 4.9KK496 pKa = 11.33SLGLQNCLFADD507 pKa = 3.64SAVSYY512 pKa = 10.6RR513 pKa = 11.84LVRR516 pKa = 11.84PGSAFHH522 pKa = 6.33PQPAGSINPPLNRR535 pKa = 11.84VRR537 pKa = 11.84PNPRR541 pKa = 11.84PRR543 pKa = 11.84LPASSLLYY551 pKa = 10.72DD552 pKa = 3.67RR553 pKa = 11.84TEE555 pKa = 3.52IMLPNMNNRR564 pKa = 11.84IVEE567 pKa = 3.99ARR569 pKa = 11.84APDD572 pKa = 4.02TLPGMTRR579 pKa = 11.84RR580 pKa = 11.84AAVNFLRR587 pKa = 11.84YY588 pKa = 8.96AQSFFGFHH596 pKa = 5.85TVDD599 pKa = 3.04SSSNALSLDD608 pKa = 4.26TIPGMTNGLLHH619 pKa = 6.79LWSPYY624 pKa = 10.56SYY626 pKa = 10.95TPFEE630 pKa = 4.16NDD632 pKa = 3.39YY633 pKa = 11.28FPDD636 pKa = 4.68PDD638 pKa = 3.68LAEE641 pKa = 4.2SRR643 pKa = 11.84HH644 pKa = 5.29YY645 pKa = 10.56FLSNLRR651 pKa = 11.84TIFGTDD657 pKa = 3.34YY658 pKa = 11.38NLVQAKK664 pKa = 9.69HH665 pKa = 6.38PYY667 pKa = 8.61EE668 pKa = 4.31ALPVVV673 pKa = 3.92
MM1 pKa = 7.81SDD3 pKa = 2.97TVLQAKK9 pKa = 8.92TNFLKK14 pKa = 10.97NKK16 pKa = 10.01SPVLSVQEE24 pKa = 4.15SKK26 pKa = 10.7RR27 pKa = 11.84PKK29 pKa = 9.63ISRR32 pKa = 11.84DD33 pKa = 3.41LMPLFQSNTAARR45 pKa = 11.84SDD47 pKa = 4.47SSNQWTIDD55 pKa = 3.26VFPNMVPITMYY66 pKa = 11.06IMLTSIAHH74 pKa = 6.28ANDD77 pKa = 2.82VDD79 pKa = 3.51HH80 pKa = 6.89RR81 pKa = 11.84TTSKK85 pKa = 10.83VSVATICMYY94 pKa = 10.53HH95 pKa = 7.74LSIVYY100 pKa = 9.77GYY102 pKa = 10.95FLLNDD107 pKa = 4.23MITRR111 pKa = 11.84ASASAHH117 pKa = 4.79ARR119 pKa = 11.84PWIEE123 pKa = 3.89TSWKK127 pKa = 10.46RR128 pKa = 11.84EE129 pKa = 3.71FADD132 pKa = 4.66FLLTLPVPEE141 pKa = 4.83FLTPFLSQFQHH152 pKa = 5.65FTTDD156 pKa = 2.65RR157 pKa = 11.84TKK159 pKa = 11.2NVFLTPSAAGFDD171 pKa = 3.75HH172 pKa = 7.12NIFFGRR178 pKa = 11.84VFPLNMFSAIHH189 pKa = 7.12DD190 pKa = 4.03CTATLPGNTPRR201 pKa = 11.84IQVLQDD207 pKa = 3.45LYY209 pKa = 11.79SRR211 pKa = 11.84VLYY214 pKa = 9.92TITAPGFTCLIPDD227 pKa = 5.29LIGVTIDD234 pKa = 3.09QTTPTTANYY243 pKa = 7.91MNSKK247 pKa = 10.1LYY249 pKa = 10.06QVYY252 pKa = 7.91TAVFNPVLFRR262 pKa = 11.84DD263 pKa = 4.41FQHH266 pKa = 6.84RR267 pKa = 11.84SSLAALSFQSPVFANNHH284 pKa = 4.01VNAYY288 pKa = 10.02DD289 pKa = 3.97LLFSASAANLRR300 pKa = 11.84EE301 pKa = 4.29LKK303 pKa = 10.65VVLQVVSTVLDD314 pKa = 4.01GKK316 pKa = 9.33VTTSGTLGDD325 pKa = 4.83FLTKK329 pKa = 10.28PSGALAIKK337 pKa = 10.15HH338 pKa = 6.07GYY340 pKa = 7.32STYY343 pKa = 11.44ALPTWSYY350 pKa = 8.77NTNATKK356 pKa = 10.53SAVYY360 pKa = 10.16SAITTLTLTSEE371 pKa = 4.19RR372 pKa = 11.84DD373 pKa = 3.3RR374 pKa = 11.84AQDD377 pKa = 2.67ICFLQRR383 pKa = 11.84PAAAFNATTEE393 pKa = 4.39VTDD396 pKa = 3.3VTYY399 pKa = 10.79AATATPNQAVQLPTGHH415 pKa = 6.38QLTRR419 pKa = 11.84HH420 pKa = 5.74FPFSLLQVPNGAEE433 pKa = 3.69PFPRR437 pKa = 11.84HH438 pKa = 6.69DD439 pKa = 4.47NDD441 pKa = 5.27DD442 pKa = 3.81LVQFSDD448 pKa = 4.81FVHH451 pKa = 6.43TAPSVLVLDD460 pKa = 4.18TDD462 pKa = 4.33GEE464 pKa = 4.52LTISAHH470 pKa = 6.48LPLLAGKK477 pKa = 9.71IIEE480 pKa = 4.27SFEE483 pKa = 4.19IDD485 pKa = 3.09GSTIEE490 pKa = 4.59MPNVDD495 pKa = 4.9KK496 pKa = 11.33SLGLQNCLFADD507 pKa = 3.64SAVSYY512 pKa = 10.6RR513 pKa = 11.84LVRR516 pKa = 11.84PGSAFHH522 pKa = 6.33PQPAGSINPPLNRR535 pKa = 11.84VRR537 pKa = 11.84PNPRR541 pKa = 11.84PRR543 pKa = 11.84LPASSLLYY551 pKa = 10.72DD552 pKa = 3.67RR553 pKa = 11.84TEE555 pKa = 3.52IMLPNMNNRR564 pKa = 11.84IVEE567 pKa = 3.99ARR569 pKa = 11.84APDD572 pKa = 4.02TLPGMTRR579 pKa = 11.84RR580 pKa = 11.84AAVNFLRR587 pKa = 11.84YY588 pKa = 8.96AQSFFGFHH596 pKa = 5.85TVDD599 pKa = 3.04SSSNALSLDD608 pKa = 4.26TIPGMTNGLLHH619 pKa = 6.79LWSPYY624 pKa = 10.56SYY626 pKa = 10.95TPFEE630 pKa = 4.16NDD632 pKa = 3.39YY633 pKa = 11.28FPDD636 pKa = 4.68PDD638 pKa = 3.68LAEE641 pKa = 4.2SRR643 pKa = 11.84HH644 pKa = 5.29YY645 pKa = 10.56FLSNLRR651 pKa = 11.84TIFGTDD657 pKa = 3.34YY658 pKa = 11.38NLVQAKK664 pKa = 9.69HH665 pKa = 6.38PYY667 pKa = 8.61EE668 pKa = 4.31ALPVVV673 pKa = 3.92
Molecular weight: 74.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1396 |
673 |
723 |
698.0 |
79.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.948 ± 1.104 | 1.074 ± 0.219 |
6.16 ± 0.34 | 3.51 ± 0.65 |
5.874 ± 0.242 | 3.51 ± 0.037 |
3.009 ± 0.024 | 5.301 ± 0.656 |
3.797 ± 0.84 | 10.172 ± 0.25 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.077 ± 0.002 | 5.372 ± 0.181 |
7.092 ± 0.125 | 3.725 ± 0.105 |
5.229 ± 0.215 | 7.235 ± 0.816 |
8.309 ± 0.695 | 5.301 ± 0.916 |
0.573 ± 0.112 | 5.731 ± 1.431 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
