
Mycoplasma gallisepticum (strain R(low / passage 15 / clone 2))
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; Mycoplasma gallisepticum; Mycoplasma gallisepticum str. R
Average proteome isoelectric point is 7.37
Get precalculated fractions of proteins
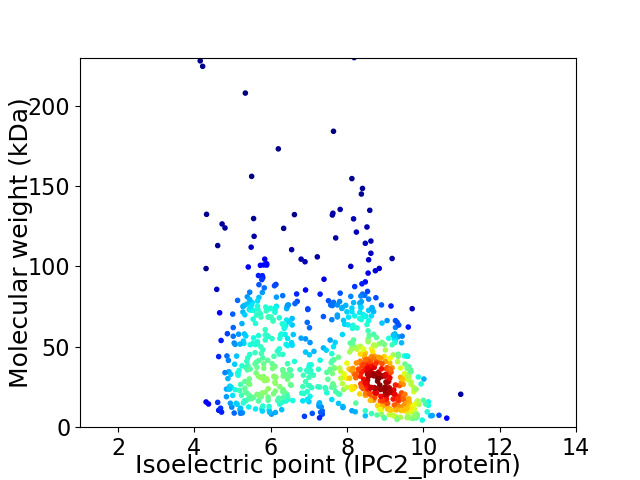
Virtual 2D-PAGE plot for 761 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q7NBG0|SYI_MYCGA Isoleucine--tRNA ligase OS=Mycoplasma gallisepticum (strain R(low / passage 15 / clone 2)) OX=710127 GN=ileS PE=3 SV=2
MM1 pKa = 8.26DD2 pKa = 4.84NNQNNFNQPGQQGFDD17 pKa = 3.05QYY19 pKa = 10.82QQQSGALVSYY29 pKa = 10.5GYY31 pKa = 9.95DD32 pKa = 3.17ANGNPVSDD40 pKa = 4.58PSLAVYY46 pKa = 9.34DD47 pKa = 4.24ANGMLINQNQYY58 pKa = 10.8DD59 pKa = 3.67QNQQQEE65 pKa = 4.14YY66 pKa = 9.14DD67 pKa = 3.32QYY69 pKa = 10.47GNPVGMLSGNVYY81 pKa = 10.34SQEE84 pKa = 3.76NDD86 pKa = 3.23PYY88 pKa = 8.68YY89 pKa = 10.26QQYY92 pKa = 9.78NQQQNQGYY100 pKa = 7.05EE101 pKa = 3.97QQYY104 pKa = 11.21DD105 pKa = 3.83EE106 pKa = 4.56YY107 pKa = 10.56GNPIGLLPGSEE118 pKa = 3.9NQQNNQQQYY127 pKa = 9.11DD128 pKa = 3.58QYY130 pKa = 10.77GNPIGMLPGGTNDD143 pKa = 3.45QAYY146 pKa = 10.43DD147 pKa = 4.11PNQMQYY153 pKa = 10.87DD154 pKa = 3.78QYY156 pKa = 10.59GNPVGMLAGVNANDD170 pKa = 3.64QGYY173 pKa = 9.85DD174 pKa = 3.86YY175 pKa = 11.62NQMQYY180 pKa = 11.14DD181 pKa = 4.1EE182 pKa = 4.67YY183 pKa = 10.41GNPVGMLPGGNANDD197 pKa = 3.28QGYY200 pKa = 9.94DD201 pKa = 3.86YY202 pKa = 11.62NQMQYY207 pKa = 11.1DD208 pKa = 4.18EE209 pKa = 4.97YY210 pKa = 11.03GNPPALYY217 pKa = 10.54DD218 pKa = 4.02NNQQDD223 pKa = 4.6YY224 pKa = 11.05YY225 pKa = 11.45GYY227 pKa = 10.49DD228 pKa = 3.01QNQQYY233 pKa = 10.85DD234 pKa = 3.41QANNQLAVVDD244 pKa = 4.29EE245 pKa = 4.96NYY247 pKa = 10.36EE248 pKa = 3.79QEE250 pKa = 4.26QQVEE254 pKa = 4.68SNEE257 pKa = 4.18EE258 pKa = 3.73PAHH261 pKa = 5.19EE262 pKa = 3.88QDD264 pKa = 5.39LRR266 pKa = 11.84EE267 pKa = 3.99FLNNNSDD274 pKa = 4.01TEE276 pKa = 4.22LVSYY280 pKa = 10.65YY281 pKa = 10.34EE282 pKa = 4.11EE283 pKa = 4.74EE284 pKa = 4.09EE285 pKa = 4.48DD286 pKa = 4.49EE287 pKa = 4.32KK288 pKa = 11.4KK289 pKa = 10.43PRR291 pKa = 11.84NKK293 pKa = 9.98KK294 pKa = 9.06KK295 pKa = 10.25QRR297 pKa = 11.84QTAQARR303 pKa = 11.84GLLPEE308 pKa = 4.95LATVNQPDD316 pKa = 3.61QTPITPHH323 pKa = 7.34LEE325 pKa = 4.24APVHH329 pKa = 5.6YY330 pKa = 10.21DD331 pKa = 3.21EE332 pKa = 6.05NEE334 pKa = 3.94EE335 pKa = 4.94LSTDD339 pKa = 4.65DD340 pKa = 5.98LSDD343 pKa = 6.31DD344 pKa = 4.1INLDD348 pKa = 3.28QNQAVHH354 pKa = 6.97HH355 pKa = 6.64DD356 pKa = 3.99AEE358 pKa = 6.21DD359 pKa = 3.65DD360 pKa = 4.16VINIPIEE367 pKa = 4.76DD368 pKa = 4.13IEE370 pKa = 4.58SALLPKK376 pKa = 10.39FEE378 pKa = 4.97EE379 pKa = 4.23IQRR382 pKa = 11.84HH383 pKa = 4.53NLEE386 pKa = 4.76EE387 pKa = 3.95IQKK390 pKa = 9.72VKK392 pKa = 11.11LEE394 pKa = 3.85AAEE397 pKa = 4.03NFKK400 pKa = 10.83VLQRR404 pKa = 11.84ANEE407 pKa = 4.0EE408 pKa = 4.15LKK410 pKa = 11.1SSNNEE415 pKa = 3.71LKK417 pKa = 8.11TTNQQLKK424 pKa = 8.71TSNEE428 pKa = 3.76ALEE431 pKa = 4.82DD432 pKa = 3.62SNKK435 pKa = 10.39RR436 pKa = 11.84IEE438 pKa = 4.44SQLQALLDD446 pKa = 4.33SINDD450 pKa = 3.26IKK452 pKa = 11.31SKK454 pKa = 10.94NNQPSQEE461 pKa = 4.21EE462 pKa = 3.86VDD464 pKa = 3.75SRR466 pKa = 11.84SKK468 pKa = 11.08LEE470 pKa = 3.75QRR472 pKa = 11.84LEE474 pKa = 3.84EE475 pKa = 4.04LAYY478 pKa = 10.01KK479 pKa = 10.42LDD481 pKa = 3.65QTKK484 pKa = 10.6EE485 pKa = 4.15IIDD488 pKa = 3.78EE489 pKa = 4.2TSEE492 pKa = 3.95SSRR495 pKa = 11.84EE496 pKa = 3.93SFQQSKK502 pKa = 10.71DD503 pKa = 3.33QLIEE507 pKa = 3.92NFEE510 pKa = 4.28KK511 pKa = 10.56KK512 pKa = 9.89IEE514 pKa = 4.1LLTEE518 pKa = 4.22KK519 pKa = 10.58LNQTQEE525 pKa = 4.26SFNQSQEE532 pKa = 4.05AKK534 pKa = 9.45QQQDD538 pKa = 2.98KK539 pKa = 11.07EE540 pKa = 4.3FAQKK544 pKa = 9.76IEE546 pKa = 4.5RR547 pKa = 11.84IIEE550 pKa = 3.96QTKK553 pKa = 9.33EE554 pKa = 4.17ANDD557 pKa = 3.6SLQNRR562 pKa = 11.84VKK564 pKa = 10.61EE565 pKa = 4.21SSLDD569 pKa = 3.66MEE571 pKa = 5.11SKK573 pKa = 10.86LEE575 pKa = 3.94NKK577 pKa = 9.99FEE579 pKa = 4.2SFADD583 pKa = 3.4KK584 pKa = 9.89LTEE587 pKa = 3.86ITSKK591 pKa = 10.7KK592 pKa = 9.49ISEE595 pKa = 4.4KK596 pKa = 8.46MTEE599 pKa = 3.88QQASKK604 pKa = 10.76KK605 pKa = 10.35EE606 pKa = 4.26EE607 pKa = 3.96IDD609 pKa = 3.6SLQSSFQKK617 pKa = 10.82ALSEE621 pKa = 4.23VVSKK625 pKa = 11.11VEE627 pKa = 3.9NYY629 pKa = 10.9ANQSQLQSQHH639 pKa = 6.2LNQTLSYY646 pKa = 10.03HH647 pKa = 4.97QQQINNSLRR656 pKa = 11.84QSAQMAQMAQMQHH669 pKa = 5.81MMPQQMLMGMNNPYY683 pKa = 10.42GFNHH687 pKa = 6.37HH688 pKa = 5.78TMMPPVHH695 pKa = 6.64QYY697 pKa = 9.57QQLPPPPPVQAPAQQLLPTINNPLQLPNEE726 pKa = 4.14NPTLFEE732 pKa = 4.8KK733 pKa = 11.01LMLANMFKK741 pKa = 9.22QTINPPQPQPQALPQPHH758 pKa = 7.1PQPQQLPPQILALPPTVVQQPNYY781 pKa = 10.26LVPQPPRR788 pKa = 11.84QPDD791 pKa = 4.02YY792 pKa = 11.1YY793 pKa = 11.06SNRR796 pKa = 11.84LNEE799 pKa = 5.01RR800 pKa = 11.84MMLDD804 pKa = 3.4DD805 pKa = 5.83AYY807 pKa = 11.52NAGYY811 pKa = 10.66DD812 pKa = 3.48EE813 pKa = 5.26AVYY816 pKa = 10.29EE817 pKa = 4.93LEE819 pKa = 4.07NQYY822 pKa = 10.67YY823 pKa = 8.58PPAYY827 pKa = 9.21EE828 pKa = 3.85YY829 pKa = 10.43PEE831 pKa = 4.15YY832 pKa = 11.16EE833 pKa = 4.23EE834 pKa = 4.28IQPSFRR840 pKa = 11.84RR841 pKa = 11.84RR842 pKa = 11.84GGRR845 pKa = 11.84AKK847 pKa = 10.39FDD849 pKa = 3.83PYY851 pKa = 11.54NNRR854 pKa = 3.4
MM1 pKa = 8.26DD2 pKa = 4.84NNQNNFNQPGQQGFDD17 pKa = 3.05QYY19 pKa = 10.82QQQSGALVSYY29 pKa = 10.5GYY31 pKa = 9.95DD32 pKa = 3.17ANGNPVSDD40 pKa = 4.58PSLAVYY46 pKa = 9.34DD47 pKa = 4.24ANGMLINQNQYY58 pKa = 10.8DD59 pKa = 3.67QNQQQEE65 pKa = 4.14YY66 pKa = 9.14DD67 pKa = 3.32QYY69 pKa = 10.47GNPVGMLSGNVYY81 pKa = 10.34SQEE84 pKa = 3.76NDD86 pKa = 3.23PYY88 pKa = 8.68YY89 pKa = 10.26QQYY92 pKa = 9.78NQQQNQGYY100 pKa = 7.05EE101 pKa = 3.97QQYY104 pKa = 11.21DD105 pKa = 3.83EE106 pKa = 4.56YY107 pKa = 10.56GNPIGLLPGSEE118 pKa = 3.9NQQNNQQQYY127 pKa = 9.11DD128 pKa = 3.58QYY130 pKa = 10.77GNPIGMLPGGTNDD143 pKa = 3.45QAYY146 pKa = 10.43DD147 pKa = 4.11PNQMQYY153 pKa = 10.87DD154 pKa = 3.78QYY156 pKa = 10.59GNPVGMLAGVNANDD170 pKa = 3.64QGYY173 pKa = 9.85DD174 pKa = 3.86YY175 pKa = 11.62NQMQYY180 pKa = 11.14DD181 pKa = 4.1EE182 pKa = 4.67YY183 pKa = 10.41GNPVGMLPGGNANDD197 pKa = 3.28QGYY200 pKa = 9.94DD201 pKa = 3.86YY202 pKa = 11.62NQMQYY207 pKa = 11.1DD208 pKa = 4.18EE209 pKa = 4.97YY210 pKa = 11.03GNPPALYY217 pKa = 10.54DD218 pKa = 4.02NNQQDD223 pKa = 4.6YY224 pKa = 11.05YY225 pKa = 11.45GYY227 pKa = 10.49DD228 pKa = 3.01QNQQYY233 pKa = 10.85DD234 pKa = 3.41QANNQLAVVDD244 pKa = 4.29EE245 pKa = 4.96NYY247 pKa = 10.36EE248 pKa = 3.79QEE250 pKa = 4.26QQVEE254 pKa = 4.68SNEE257 pKa = 4.18EE258 pKa = 3.73PAHH261 pKa = 5.19EE262 pKa = 3.88QDD264 pKa = 5.39LRR266 pKa = 11.84EE267 pKa = 3.99FLNNNSDD274 pKa = 4.01TEE276 pKa = 4.22LVSYY280 pKa = 10.65YY281 pKa = 10.34EE282 pKa = 4.11EE283 pKa = 4.74EE284 pKa = 4.09EE285 pKa = 4.48DD286 pKa = 4.49EE287 pKa = 4.32KK288 pKa = 11.4KK289 pKa = 10.43PRR291 pKa = 11.84NKK293 pKa = 9.98KK294 pKa = 9.06KK295 pKa = 10.25QRR297 pKa = 11.84QTAQARR303 pKa = 11.84GLLPEE308 pKa = 4.95LATVNQPDD316 pKa = 3.61QTPITPHH323 pKa = 7.34LEE325 pKa = 4.24APVHH329 pKa = 5.6YY330 pKa = 10.21DD331 pKa = 3.21EE332 pKa = 6.05NEE334 pKa = 3.94EE335 pKa = 4.94LSTDD339 pKa = 4.65DD340 pKa = 5.98LSDD343 pKa = 6.31DD344 pKa = 4.1INLDD348 pKa = 3.28QNQAVHH354 pKa = 6.97HH355 pKa = 6.64DD356 pKa = 3.99AEE358 pKa = 6.21DD359 pKa = 3.65DD360 pKa = 4.16VINIPIEE367 pKa = 4.76DD368 pKa = 4.13IEE370 pKa = 4.58SALLPKK376 pKa = 10.39FEE378 pKa = 4.97EE379 pKa = 4.23IQRR382 pKa = 11.84HH383 pKa = 4.53NLEE386 pKa = 4.76EE387 pKa = 3.95IQKK390 pKa = 9.72VKK392 pKa = 11.11LEE394 pKa = 3.85AAEE397 pKa = 4.03NFKK400 pKa = 10.83VLQRR404 pKa = 11.84ANEE407 pKa = 4.0EE408 pKa = 4.15LKK410 pKa = 11.1SSNNEE415 pKa = 3.71LKK417 pKa = 8.11TTNQQLKK424 pKa = 8.71TSNEE428 pKa = 3.76ALEE431 pKa = 4.82DD432 pKa = 3.62SNKK435 pKa = 10.39RR436 pKa = 11.84IEE438 pKa = 4.44SQLQALLDD446 pKa = 4.33SINDD450 pKa = 3.26IKK452 pKa = 11.31SKK454 pKa = 10.94NNQPSQEE461 pKa = 4.21EE462 pKa = 3.86VDD464 pKa = 3.75SRR466 pKa = 11.84SKK468 pKa = 11.08LEE470 pKa = 3.75QRR472 pKa = 11.84LEE474 pKa = 3.84EE475 pKa = 4.04LAYY478 pKa = 10.01KK479 pKa = 10.42LDD481 pKa = 3.65QTKK484 pKa = 10.6EE485 pKa = 4.15IIDD488 pKa = 3.78EE489 pKa = 4.2TSEE492 pKa = 3.95SSRR495 pKa = 11.84EE496 pKa = 3.93SFQQSKK502 pKa = 10.71DD503 pKa = 3.33QLIEE507 pKa = 3.92NFEE510 pKa = 4.28KK511 pKa = 10.56KK512 pKa = 9.89IEE514 pKa = 4.1LLTEE518 pKa = 4.22KK519 pKa = 10.58LNQTQEE525 pKa = 4.26SFNQSQEE532 pKa = 4.05AKK534 pKa = 9.45QQQDD538 pKa = 2.98KK539 pKa = 11.07EE540 pKa = 4.3FAQKK544 pKa = 9.76IEE546 pKa = 4.5RR547 pKa = 11.84IIEE550 pKa = 3.96QTKK553 pKa = 9.33EE554 pKa = 4.17ANDD557 pKa = 3.6SLQNRR562 pKa = 11.84VKK564 pKa = 10.61EE565 pKa = 4.21SSLDD569 pKa = 3.66MEE571 pKa = 5.11SKK573 pKa = 10.86LEE575 pKa = 3.94NKK577 pKa = 9.99FEE579 pKa = 4.2SFADD583 pKa = 3.4KK584 pKa = 9.89LTEE587 pKa = 3.86ITSKK591 pKa = 10.7KK592 pKa = 9.49ISEE595 pKa = 4.4KK596 pKa = 8.46MTEE599 pKa = 3.88QQASKK604 pKa = 10.76KK605 pKa = 10.35EE606 pKa = 4.26EE607 pKa = 3.96IDD609 pKa = 3.6SLQSSFQKK617 pKa = 10.82ALSEE621 pKa = 4.23VVSKK625 pKa = 11.11VEE627 pKa = 3.9NYY629 pKa = 10.9ANQSQLQSQHH639 pKa = 6.2LNQTLSYY646 pKa = 10.03HH647 pKa = 4.97QQQINNSLRR656 pKa = 11.84QSAQMAQMAQMQHH669 pKa = 5.81MMPQQMLMGMNNPYY683 pKa = 10.42GFNHH687 pKa = 6.37HH688 pKa = 5.78TMMPPVHH695 pKa = 6.64QYY697 pKa = 9.57QQLPPPPPVQAPAQQLLPTINNPLQLPNEE726 pKa = 4.14NPTLFEE732 pKa = 4.8KK733 pKa = 11.01LMLANMFKK741 pKa = 9.22QTINPPQPQPQALPQPHH758 pKa = 7.1PQPQQLPPQILALPPTVVQQPNYY781 pKa = 10.26LVPQPPRR788 pKa = 11.84QPDD791 pKa = 4.02YY792 pKa = 11.1YY793 pKa = 11.06SNRR796 pKa = 11.84LNEE799 pKa = 5.01RR800 pKa = 11.84MMLDD804 pKa = 3.4DD805 pKa = 5.83AYY807 pKa = 11.52NAGYY811 pKa = 10.66DD812 pKa = 3.48EE813 pKa = 5.26AVYY816 pKa = 10.29EE817 pKa = 4.93LEE819 pKa = 4.07NQYY822 pKa = 10.67YY823 pKa = 8.58PPAYY827 pKa = 9.21EE828 pKa = 3.85YY829 pKa = 10.43PEE831 pKa = 4.15YY832 pKa = 11.16EE833 pKa = 4.23EE834 pKa = 4.28IQPSFRR840 pKa = 11.84RR841 pKa = 11.84RR842 pKa = 11.84GGRR845 pKa = 11.84AKK847 pKa = 10.39FDD849 pKa = 3.83PYY851 pKa = 11.54NNRR854 pKa = 3.4
Molecular weight: 98.69 kDa
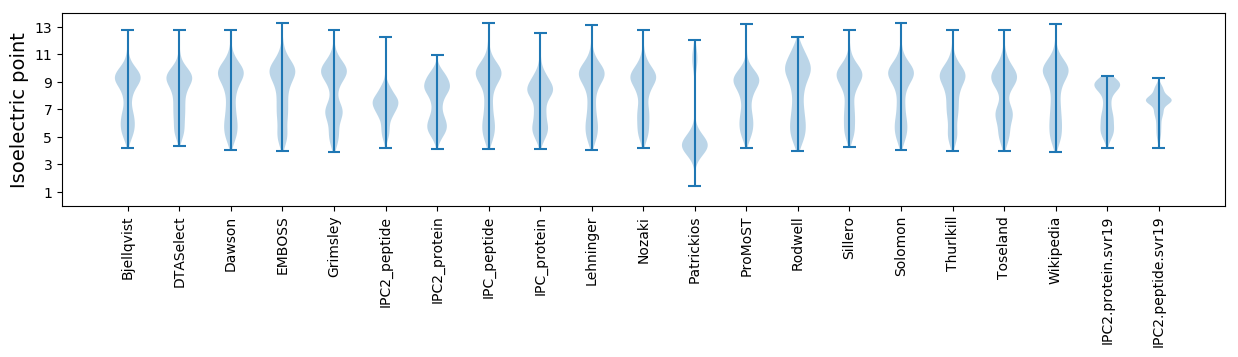
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8WJY3|F8WJY3_MYCGA High affinity transport system protein OS=Mycoplasma gallisepticum (strain R(low / passage 15 / clone 2)) OX=710127 GN=hatA PE=4 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84PAPSPMGSPKK12 pKa = 10.69LLGPNQAGHH21 pKa = 6.37PQHH24 pKa = 6.8GPRR27 pKa = 11.84PMNAHH32 pKa = 7.34PGQPRR37 pKa = 11.84PQQAGPRR44 pKa = 11.84PMGAGGSNQPRR55 pKa = 11.84PMPNGLQNPQGPRR68 pKa = 11.84PMNPQGDD75 pKa = 4.49PRR77 pKa = 11.84PQPAGVRR84 pKa = 11.84PNSPQNSQPRR94 pKa = 11.84PMPNKK99 pKa = 8.94PQGPRR104 pKa = 11.84PMGAPNPQPGPQQAGPRR121 pKa = 11.84PMGVGGSNQPRR132 pKa = 11.84PMPNGLQNPQGPRR145 pKa = 11.84PMNPQGDD152 pKa = 4.49PRR154 pKa = 11.84PQPAGVRR161 pKa = 11.84PNSPQANQPGRR172 pKa = 11.84RR173 pKa = 11.84PTPNNPQGPRR183 pKa = 11.84PMGPRR188 pKa = 11.84PNGGPNRR195 pKa = 11.84AA196 pKa = 3.81
MM1 pKa = 7.68RR2 pKa = 11.84PAPSPMGSPKK12 pKa = 10.69LLGPNQAGHH21 pKa = 6.37PQHH24 pKa = 6.8GPRR27 pKa = 11.84PMNAHH32 pKa = 7.34PGQPRR37 pKa = 11.84PQQAGPRR44 pKa = 11.84PMGAGGSNQPRR55 pKa = 11.84PMPNGLQNPQGPRR68 pKa = 11.84PMNPQGDD75 pKa = 4.49PRR77 pKa = 11.84PQPAGVRR84 pKa = 11.84PNSPQNSQPRR94 pKa = 11.84PMPNKK99 pKa = 8.94PQGPRR104 pKa = 11.84PMGAPNPQPGPQQAGPRR121 pKa = 11.84PMGVGGSNQPRR132 pKa = 11.84PMPNGLQNPQGPRR145 pKa = 11.84PMNPQGDD152 pKa = 4.49PRR154 pKa = 11.84PQPAGVRR161 pKa = 11.84PNSPQANQPGRR172 pKa = 11.84RR173 pKa = 11.84PTPNNPQGPRR183 pKa = 11.84PMGPRR188 pKa = 11.84PNGGPNRR195 pKa = 11.84AA196 pKa = 3.81
Molecular weight: 20.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
295319 |
37 |
1969 |
388.1 |
44.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.855 ± 0.087 | 0.519 ± 0.021 |
5.679 ± 0.068 | 5.605 ± 0.146 |
4.925 ± 0.081 | 4.675 ± 0.082 |
1.437 ± 0.039 | 8.244 ± 0.118 |
8.903 ± 0.108 | 9.789 ± 0.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.686 ± 0.036 | 8.033 ± 0.105 |
3.255 ± 0.062 | 4.213 ± 0.095 |
3.159 ± 0.049 | 7.119 ± 0.083 |
5.828 ± 0.109 | 5.912 ± 0.068 |
0.869 ± 0.024 | 4.293 ± 0.061 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
