
Bemisia-associated genomovirus AdO
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus bemta1
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
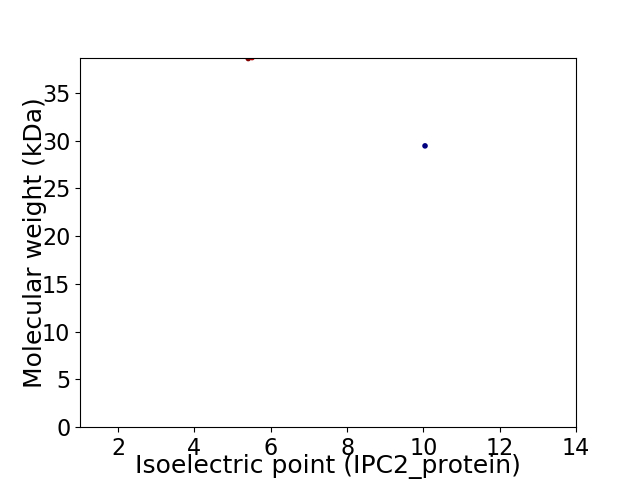
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2X292|A0A1Z2X292_9VIRU Putative coat protein OS=Bemisia-associated genomovirus AdO OX=1986478 GN=CP PE=4 SV=1
MM1 pKa = 7.75PSFVHH6 pKa = 6.39NYY8 pKa = 9.2RR9 pKa = 11.84YY10 pKa = 10.74ALVTYY15 pKa = 9.62SQYY18 pKa = 11.76GDD20 pKa = 3.79LDD22 pKa = 3.28PWRR25 pKa = 11.84VMEE28 pKa = 4.64RR29 pKa = 11.84FSSLGAEE36 pKa = 4.14CIVARR41 pKa = 11.84EE42 pKa = 4.01HH43 pKa = 7.16HH44 pKa = 6.88EE45 pKa = 4.44DD46 pKa = 3.61GGLHH50 pKa = 5.69LHH52 pKa = 6.24VFADD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.87ILDD70 pKa = 3.06VDD72 pKa = 3.87GRR74 pKa = 11.84HH75 pKa = 6.37ANVEE79 pKa = 4.17PSAGTPEE86 pKa = 3.94KK87 pKa = 10.83GYY89 pKa = 10.94DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.83DD95 pKa = 3.47GDD97 pKa = 4.3VVCGGLARR105 pKa = 11.84PEE107 pKa = 3.94PSAVGDD113 pKa = 3.9GPTAAKK119 pKa = 8.46WARR122 pKa = 11.84ITSAEE127 pKa = 4.15TRR129 pKa = 11.84SEE131 pKa = 3.97FWKK134 pKa = 10.52LCHH137 pKa = 6.51EE138 pKa = 5.45LDD140 pKa = 4.41PKK142 pKa = 10.92AAACNFSALSKK153 pKa = 8.92YY154 pKa = 10.36CDD156 pKa = 2.81WRR158 pKa = 11.84FAEE161 pKa = 5.18DD162 pKa = 3.76PPEE165 pKa = 4.16YY166 pKa = 10.48EE167 pKa = 3.29HH168 pKa = 7.62DD169 pKa = 3.67GRR171 pKa = 11.84IKK173 pKa = 10.38FVPGDD178 pKa = 3.4GDD180 pKa = 4.83GRR182 pKa = 11.84DD183 pKa = 3.51DD184 pKa = 3.48WVSQSGIRR192 pKa = 11.84LDD194 pKa = 3.93EE195 pKa = 4.75PFLGMWVFAPNRR207 pKa = 11.84VKK209 pKa = 10.93SLVLYY214 pKa = 10.75GGTRR218 pKa = 11.84TGKK221 pKa = 5.93TTWARR226 pKa = 11.84SLGSHH231 pKa = 6.56VYY233 pKa = 10.64CIGLVSGTEE242 pKa = 4.09CARR245 pKa = 11.84GLDD248 pKa = 3.23AKK250 pKa = 10.93YY251 pKa = 10.81AVFDD255 pKa = 5.03DD256 pKa = 3.77IRR258 pKa = 11.84GGLAFFHH265 pKa = 7.2GYY267 pKa = 9.97KK268 pKa = 9.68EE269 pKa = 3.96WLGAQPHH276 pKa = 5.31VSIKK280 pKa = 9.68QLYY283 pKa = 8.38RR284 pKa = 11.84EE285 pKa = 4.38PYY287 pKa = 7.41YY288 pKa = 10.35MRR290 pKa = 11.84WGKK293 pKa = 7.47PTIWVCNTDD302 pKa = 3.55PRR304 pKa = 11.84LDD306 pKa = 4.29AYY308 pKa = 9.24PQNARR313 pKa = 11.84PDD315 pKa = 4.03FEE317 pKa = 4.46WMEE320 pKa = 4.29GNCTFIEE327 pKa = 4.49VTDD330 pKa = 3.95SLIEE334 pKa = 4.57SISHH338 pKa = 6.9ANTDD342 pKa = 3.19
MM1 pKa = 7.75PSFVHH6 pKa = 6.39NYY8 pKa = 9.2RR9 pKa = 11.84YY10 pKa = 10.74ALVTYY15 pKa = 9.62SQYY18 pKa = 11.76GDD20 pKa = 3.79LDD22 pKa = 3.28PWRR25 pKa = 11.84VMEE28 pKa = 4.64RR29 pKa = 11.84FSSLGAEE36 pKa = 4.14CIVARR41 pKa = 11.84EE42 pKa = 4.01HH43 pKa = 7.16HH44 pKa = 6.88EE45 pKa = 4.44DD46 pKa = 3.61GGLHH50 pKa = 5.69LHH52 pKa = 6.24VFADD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.87ILDD70 pKa = 3.06VDD72 pKa = 3.87GRR74 pKa = 11.84HH75 pKa = 6.37ANVEE79 pKa = 4.17PSAGTPEE86 pKa = 3.94KK87 pKa = 10.83GYY89 pKa = 10.94DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.83DD95 pKa = 3.47GDD97 pKa = 4.3VVCGGLARR105 pKa = 11.84PEE107 pKa = 3.94PSAVGDD113 pKa = 3.9GPTAAKK119 pKa = 8.46WARR122 pKa = 11.84ITSAEE127 pKa = 4.15TRR129 pKa = 11.84SEE131 pKa = 3.97FWKK134 pKa = 10.52LCHH137 pKa = 6.51EE138 pKa = 5.45LDD140 pKa = 4.41PKK142 pKa = 10.92AAACNFSALSKK153 pKa = 8.92YY154 pKa = 10.36CDD156 pKa = 2.81WRR158 pKa = 11.84FAEE161 pKa = 5.18DD162 pKa = 3.76PPEE165 pKa = 4.16YY166 pKa = 10.48EE167 pKa = 3.29HH168 pKa = 7.62DD169 pKa = 3.67GRR171 pKa = 11.84IKK173 pKa = 10.38FVPGDD178 pKa = 3.4GDD180 pKa = 4.83GRR182 pKa = 11.84DD183 pKa = 3.51DD184 pKa = 3.48WVSQSGIRR192 pKa = 11.84LDD194 pKa = 3.93EE195 pKa = 4.75PFLGMWVFAPNRR207 pKa = 11.84VKK209 pKa = 10.93SLVLYY214 pKa = 10.75GGTRR218 pKa = 11.84TGKK221 pKa = 5.93TTWARR226 pKa = 11.84SLGSHH231 pKa = 6.56VYY233 pKa = 10.64CIGLVSGTEE242 pKa = 4.09CARR245 pKa = 11.84GLDD248 pKa = 3.23AKK250 pKa = 10.93YY251 pKa = 10.81AVFDD255 pKa = 5.03DD256 pKa = 3.77IRR258 pKa = 11.84GGLAFFHH265 pKa = 7.2GYY267 pKa = 9.97KK268 pKa = 9.68EE269 pKa = 3.96WLGAQPHH276 pKa = 5.31VSIKK280 pKa = 9.68QLYY283 pKa = 8.38RR284 pKa = 11.84EE285 pKa = 4.38PYY287 pKa = 7.41YY288 pKa = 10.35MRR290 pKa = 11.84WGKK293 pKa = 7.47PTIWVCNTDD302 pKa = 3.55PRR304 pKa = 11.84LDD306 pKa = 4.29AYY308 pKa = 9.24PQNARR313 pKa = 11.84PDD315 pKa = 4.03FEE317 pKa = 4.46WMEE320 pKa = 4.29GNCTFIEE327 pKa = 4.49VTDD330 pKa = 3.95SLIEE334 pKa = 4.57SISHH338 pKa = 6.9ANTDD342 pKa = 3.19
Molecular weight: 38.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2X292|A0A1Z2X292_9VIRU Putative coat protein OS=Bemisia-associated genomovirus AdO OX=1986478 GN=CP PE=4 SV=1
MM1 pKa = 7.65ALRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84TTRR12 pKa = 11.84KK13 pKa = 9.58RR14 pKa = 11.84PARR17 pKa = 11.84RR18 pKa = 11.84SGGTTSRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 5.04PTTRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 7.54PRR37 pKa = 11.84KK38 pKa = 9.1MSNKK42 pKa = 9.88RR43 pKa = 11.84ILNLTARR50 pKa = 11.84KK51 pKa = 9.74KK52 pKa = 10.39RR53 pKa = 11.84DD54 pKa = 3.14TMQSFTNLSAAGGTATFAAQPAFLYY79 pKa = 10.61GGRR82 pKa = 11.84TYY84 pKa = 10.72ILPWIATARR93 pKa = 11.84DD94 pKa = 3.26LSSNVNAPNTIAQEE108 pKa = 4.28SARR111 pKa = 11.84TATVCYY117 pKa = 9.75MRR119 pKa = 11.84GLKK122 pKa = 9.83EE123 pKa = 4.19RR124 pKa = 11.84IQIQTSSGIPWQWRR138 pKa = 11.84RR139 pKa = 11.84ICFTVKK145 pKa = 10.68GNDD148 pKa = 3.5LLIGVNGDD156 pKa = 3.8GDD158 pKa = 3.9RR159 pKa = 11.84QPLKK163 pKa = 9.3EE164 pKa = 4.18TTFGFMRR171 pKa = 11.84NVVDD175 pKa = 3.91WNDD178 pKa = 3.42KK179 pKa = 10.63ASAAIPLTEE188 pKa = 4.76ALFKK192 pKa = 10.78GQRR195 pKa = 11.84NTDD198 pKa = 2.83WRR200 pKa = 11.84SLLNAPVDD208 pKa = 3.86TRR210 pKa = 11.84RR211 pKa = 11.84VTLKK215 pKa = 10.19YY216 pKa = 10.46DD217 pKa = 3.12KK218 pKa = 10.81TRR220 pKa = 11.84IISSGNSNGVLRR232 pKa = 11.84NFNLWHH238 pKa = 6.72PMNKK242 pKa = 9.55NLVYY246 pKa = 10.83GDD248 pKa = 4.69DD249 pKa = 3.38EE250 pKa = 4.78GGNYY254 pKa = 9.76DD255 pKa = 4.02VEE257 pKa = 4.65YY258 pKa = 11.15
MM1 pKa = 7.65ALRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84TTRR12 pKa = 11.84KK13 pKa = 9.58RR14 pKa = 11.84PARR17 pKa = 11.84RR18 pKa = 11.84SGGTTSRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 5.04PTTRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 7.54PRR37 pKa = 11.84KK38 pKa = 9.1MSNKK42 pKa = 9.88RR43 pKa = 11.84ILNLTARR50 pKa = 11.84KK51 pKa = 9.74KK52 pKa = 10.39RR53 pKa = 11.84DD54 pKa = 3.14TMQSFTNLSAAGGTATFAAQPAFLYY79 pKa = 10.61GGRR82 pKa = 11.84TYY84 pKa = 10.72ILPWIATARR93 pKa = 11.84DD94 pKa = 3.26LSSNVNAPNTIAQEE108 pKa = 4.28SARR111 pKa = 11.84TATVCYY117 pKa = 9.75MRR119 pKa = 11.84GLKK122 pKa = 9.83EE123 pKa = 4.19RR124 pKa = 11.84IQIQTSSGIPWQWRR138 pKa = 11.84RR139 pKa = 11.84ICFTVKK145 pKa = 10.68GNDD148 pKa = 3.5LLIGVNGDD156 pKa = 3.8GDD158 pKa = 3.9RR159 pKa = 11.84QPLKK163 pKa = 9.3EE164 pKa = 4.18TTFGFMRR171 pKa = 11.84NVVDD175 pKa = 3.91WNDD178 pKa = 3.42KK179 pKa = 10.63ASAAIPLTEE188 pKa = 4.76ALFKK192 pKa = 10.78GQRR195 pKa = 11.84NTDD198 pKa = 2.83WRR200 pKa = 11.84SLLNAPVDD208 pKa = 3.86TRR210 pKa = 11.84RR211 pKa = 11.84VTLKK215 pKa = 10.19YY216 pKa = 10.46DD217 pKa = 3.12KK218 pKa = 10.81TRR220 pKa = 11.84IISSGNSNGVLRR232 pKa = 11.84NFNLWHH238 pKa = 6.72PMNKK242 pKa = 9.55NLVYY246 pKa = 10.83GDD248 pKa = 4.69DD249 pKa = 3.38EE250 pKa = 4.78GGNYY254 pKa = 9.76DD255 pKa = 4.02VEE257 pKa = 4.65YY258 pKa = 11.15
Molecular weight: 29.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
600 |
258 |
342 |
300.0 |
34.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.0 ± 0.146 | 1.833 ± 0.625 |
6.833 ± 1.06 | 4.5 ± 1.284 |
4.0 ± 0.531 | 8.5 ± 0.671 |
2.167 ± 1.051 | 4.167 ± 0.286 |
4.5 ± 0.318 | 6.833 ± 0.314 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.291 | 4.5 ± 1.691 |
5.167 ± 0.304 | 2.167 ± 0.552 |
9.833 ± 1.975 | 6.0 ± 0.11 |
6.833 ± 1.687 | 5.333 ± 0.632 |
2.833 ± 0.3 | 4.167 ± 0.401 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
